Abstract
The FireDB database is a databank for functional information relating to proteins with known structures. It contains the most comprehensive and detailed repository of known functionally important residues, bringing together both ligand binding and catalytic residues in one site. The platform integrates biologically relevant data filtered from the close atomic contacts in Protein Data Bank crystal structures and reliably annotated catalytic residues from the Catalytic Site Atlas. The interface allows users to make queries by protein, ligand or keyword. Relevant biologically important residues are displayed in a simple and easy to read manner that allows users to assess binding site similarity across homologous proteins. Binding site residue variations can also be viewed with molecular visualization tools. The database is available at http://firedb.bioinfo.cnio.es
INTRODUCTION
For many years the Protein Data Bank (PDB) (1) has been the primary source of information about biological macromolecules (2). The PDB began in 1971 with seven structures and in recent years has seen dramatic growth. This acceleration in the deposition rate, a consequence of structural genomics initiatives (3), means that the PDB now has over 38 000 entries. The increase in the number and complexity of protein structures in the PDB highlights the importance of creating new data mining and analytical tools capable of dealing with large amounts of structural information.
Although the increase in structural data means that the structural space is being covered, many of the structures generated by structural genomics initiatives have unknown function (4). Functional space is generally regarded as being broader than structural space (5), so this lack of functional annotation is a real problem, and as an issue it is only just starting to be addressed by the structural genomic initiatives (6).
A variety of functional analysis tools already exist. For example HIC-Up (7) and PDBsum (8) are web retrieval tools designed to allow navigation across complexes with different compounds or ligands and the Ligand Depot database (9) allows characterization of ligands according to chemical and geometrical characteristics. RELIBASE (10) allows binding sites to be studied according to sequence and secondary structure similarity and in LigBase (11) binding sites are aligned with related sequence and structures. The Protein Ligand Database (PLD) (12) is a repository of protein–ligand complexes and includes energy calculations and ligand similarities, but only 485 complexes are stored in the database. PDB-ligand (13) allows comparisons between structurally similar binding sites from proteins binding the same ligand, although cases where the same residues bind different ligand analogs are not addressed.
Information on functionally important residues can be obtained from a range of sources. There is catalytic site information within the actual PDB files, although the data is not uniformly maintained and text mining is necessary to classify the residues according to their function, something that is done in the database PDBsite (14). The most important data source for catalytic sites is the Catalytic Site Atlas [CSA, Thornton et al. (15)], a curated database of catalytic sites from the PDB as well as from the literature related to structures. They also explore catalytic site evolution in homologous families by comparing sequence identity and RMSD (16).
Probably the biggest source of functionally important residues comes from close atomic contacts between protein residues and small ligands. There are several databases with different levels of organization [8, 9, 10, 12 and 13]. The number of non-biological ligands in PDB structures is a major issue when detecting ligands through protein-ligand atomic contacts, something that is especially true for small inorganic molecules and ions.
This work aims to resolve issues associated with the currently available sources of functionally important residues, firstly by integrating all the available sources into a single database and secondly by filtering and validating the ligands involved in protein-ligand atomic contacts.
CONTENTS AND METHODS
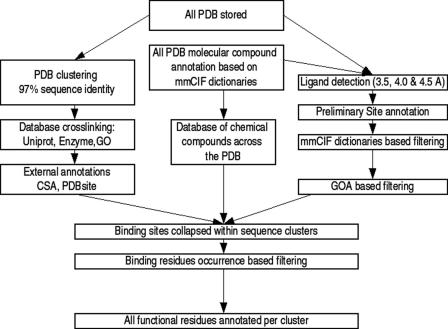
PDB sequences are extracted directly from the co-ordinates file and modified amino acids are translated to their parent standard one letter code. The main entity in FireDB is the master sequence. The master sequences are generated by clustering all PDB chains at 97% sequence identity using the program CD-HIT (17) and extracting a consensus sequence from the multiple sequence alignments built with T-coffee (18) or muscle (19) for each of the clusters (Figure 1). In the PDB, at 97% sequence identity the main differences between sequences come from structural gaps and mutations. The master sequence is the nexus between chains from the same cluster. Associated with the master sequence is an indication of residue conservation. Residue conservation is calculated for the master sequence family via SQUARE (20), a method that calculates a profile-based measure of per residue reliability.
Figure 1.
FireDB flowchart. The organization allows two search modes—molecular compounds and proteins.
FireDB in numbers
The version of FireDB presented here contains the full PDB of 14 July 2006. 76 504 protein chains (35 496 unique sequences) are processed into 15 777 master sequences. Redundancy is particularly asymmetric in the PDB; 5700 chains are represented only once while the biggest cluster, human haemoglobin sub-units alpha and beta, are represented by more than 300 chains.
Sites are found in 43 413 chains and collapsed into 8153 of the master sequences. Table 1. shows the frequency of sites and molecular compounds in PDB in different classes. Such classes are derived from the mmCIF-format components.cif file obtained from the PDB. HETAI is the most common ligand class, it includes 78 different ions and small inorganic molecules and is present in 48 718 sites. The second most common category is HETAIN (48 209 sites), a heterogeneous class containing over 4914 different compounds such as inhibitors and non-canonical biological molecules.
Table 1.
Content of FireDB, contact calculations at 4.0 Angstroms distance. The data are collected by scanning the full PDB, biological relevance of binding sites was not assessed at this point
| CLASS (mmCIF) | Description | Number of sites | Number of molecular compounds |
|---|---|---|---|
| HETAI | Ions | 48 718 | 78 |
| HETAIN | Inhibitors, non-canonical biological molecules, etc | 48 209 | 4914 |
| ATOMS | Saccharide and derivates | 14 778 | 240 |
| ATOMN | Nucleotides and derivates | 1058 | 112 |
| ATOMP | Amino acids and derivates | 906 | 97 |
| HETAC | Co-enzymes | 832 | 23 |
| HETIC | Water coordinated ions | 642 | 29 |
| HETAD | Drugs | 118 | 46 |
| Total | 115 261 | 5539 |
External data integration
The CSA provides information about active sites extracted from the literature and the PDB and extended by homology with PSI-BLAST (21) searches. The whole CSA is integrated into FireDB and displayed in tables and in visualization tools with external links back to the CSA.
MSD (22) aims to manage, collect and distribute macromolecular information. The cross-linking platform provides EC enzyme numbers (23), as well as Uniprot (24) primary accession numbers to PDB entries; both are included in FireDB.
GOA-PDB (25) provides GO term annotations for any PDB chain from a variety of sources; these terms are also added to FireDB. GO terms can be used to evaluate whether a given ligand present in the co-ordinates file is biologically relevant, as shown later.
The mmCIF (26) is a data exchange format for the Crystallographic Information Framework. The components.cif file shows the correspondence between PDB compounds and mmCIF dictionaries and it is updated when new compounds appear in PDB. It includes ligand features as formulae, molecular weights, etc. as well as classifications (Table 1) of compounds that make it possible to infer in many cases the role played by a given ligand. All this data are also integrated in FireDB.
Scanning the PDB: Ligand search, filtering and characterising
All atom contacts between proteins and heteroatoms are calculated for three distance cut-offs (3.5, 4.0 and 4.5 Å). In order to collect the most reliable set of contacting residues we prefiltered several molecular species: solvent molecules, non-biological ions and heavy atoms. FireDB is oriented towards small molecule ligands, so interactions with proteins, DNA and RNA are not considered and large ligands where the number of ligand atoms is 2/3 or greater than the number of protein atoms, such as the photosystems where multiple pigments are bound up into protein-ligand conglomerates, are also rejected. A perl script runs for any PDB file, a version that runs via a web interface is available at http://firedb.bioinfo.cnio.es/Php/Contact.php.
Small inorganic molecules and ions yield most of the non-biological binding sites, and it is often difficult to decide whether the bound ligand has biological relevance. For these cases we have developed a library cross-linking GO terms with ligand mmCIF codes. Term GO:0008270 (zinc ion binding) is related to Zinc atom code ‘ZN’, GO:0005509 (calcium ion binding) is related to Calcium code ‘CA’ and so on. The co-occurrence of a GOA term and the presence of the related ligand in a given PDB chain improves confidence. The mmCIF_2_GO library is being extended to the rest of the ligands where a related GO term exists.
Collapsing binding sites
One advantage of collapsing the structures into master sequences is that the redundancy and complexity of the database is reduced. Another major utility of the collapsed sequences is that they give the user the ability to compare binding sites within nearly identical proteins, something that is possible because all binding residues are mapped onto master sequences. Binding sites may be occupied by identical or similar ligands, and the tabular representation of the residues in contact with the ligand can highlight the flexibility of those regions. It is possible to assess the analogy of binding sites by comparing the residues in each sequence that bind each ligand. Within the same master sequence all ligands considered to be binding analogously can be viewed in the same multiple alignment, two sites A and B are considered to be analogs when 60% of the binding residues in A coincide with B and vice versa.
As part of the collapsing process, residues are given an occupancy score. Occupancy is the frequency with which the equivalent residue is in contact with a ligand in each of the sequences collapsed into the master sequence. Figure 2a shows the consensus binding residues for a number of analogous ligands from the same master sequence and Figure 2b shows the expanded version of one of the analogous sites. Here the colour scheme represents the percentage of occurrence of each residue in the master sequence as shown in Figure 3. This diagram shows that even when the master sequence is generated from many structures, the consensus residues of the binding site maintain a high percentage of occupancy, showing that the collapsing method works fine even for the most difficult cases.
Figure 2.
Information retrieved for the family of sequence collapsed around 1tcoC. (a) a representation of the analogous sites collapsed into the master sequence for the FK506 bidning proteins. The sites are ordered by ligand occupancy and are annotated with information from the Catalytic Site Atlas. (b) an expanded view of the binding residues from the sequences that bind the ligand analog RAP from the sequences collapsed into the FK506 binding protein master sequence; the colour scheme of each residue depends on the percentage of occupancy of each residue. (c) Jmol representations of four cases from this family, representing (i) the FK506 binding site for 1tcoC, (ii) the FK506 binding site for 1bkf, (iii) the RAP (Rapamycin) binding site for 1bkf and (iv) the RAP binding site for 1fapA. The backbone and ball and stick for protein, Van der Waals representation for the ligands. The residue composition for all four binding sites is similar, but different.
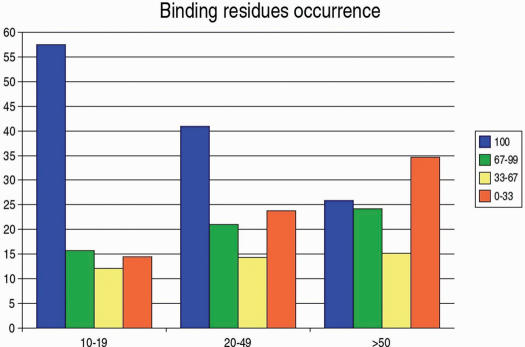
Figure 3.
A breakdown of binding residue occupancy. Binding residues are grouped into three bins depending on the number of sequences with binding sites that are collapsed into each of the master sequences. These bins were for 10–19 sequences (a total of 14, 594 binding residues), 20–49 sequences (7536 residues) and greater than 49 sequences (1937 residues). The number of collapsed sequences used for the bins is shown in the x-axis. Residues were also clustered into bins depending on the occupancy in binding sites. Occupancy for each residue in a master sequence is defined as the percentage of collapsed sequences in which each equivalent residue is in contact with the ligand at 4A. Occupancy is shown in the legend and the percentage of residues at each of the occupancy bins is shown in the y-axis. Even for the group with 50 or more structures collapsed into the master sequence, 25% of binding residues bind the ligand in every single one of the collapsed sequences and another 25% fall in the 67–99% range of occupancy percentages.
Functionality
Users of FireDB will be able to retrieve annotated site residues simply by entering the PDB code, Uniprot primary accession numbers or simple keywords related to the structure they are interested in. It will also be possible to refine queries of ligand three-letter code and keywords. The information retrieved will include the type of site, a chemical description of the ligand, the list of chains that bind the required ligand and the residues involved.
The way the data is structured allows comparison between chains that contain the same binding site, highlighting the flexibility of those regions. Information is displayed in tables and molecular visualization with the Java applet Jmol is available for this purpose.
FireDB is being built in an updatable way: new sites and new site types will be added from the weekly PDB updates. In this way FireDB will be a valuable tool for both researchers that are looking for information about individual targets and for those who wish to obtain data for broader analyses.
FireDB has collaborated in the GeneFun project (http://www.genefun.org), a project which aims to assess protein database anntation reliability and incorporate higher-level features into functional annotations. We have annotated 209 Structural Genomics targets in collaboration with other groups. The results can be browsed at http://firedb.bioinfo.cnio.es/GeneFun/results.html.
FK506 BINDING PROTEIN, A PRACTICAL CASE
FK506 binding protein catalyses the cis–trans isomerization of proline imidic peptide bonds in oligopeptides and may play a role in the modulation of ryanodine receptor isoform-1 (RYR-1), a component of the calcium release channel of skeletal muscle sarcoplasmic reticulum. It is sterically inhibited by both FK506 (FK5) and rapamycin (RAP).
There are multiple 52 versions of this protein in the PDB (at 97% sequence identity), of which 75% (39) bind a ligand in the inhibitor binding site (Figure 2a). The residue occupancy highlighted in different colours in Figure 2b suggests which residues are essential for binding in this case. Val55, Ile56, Trp59, Tyr82 and Phe99 form the essential hydrophobic environment while Arg42 and Gln53 bind selectively to FK5 and RAP respectively.
Molecule visualization is possible in FireDB; in Figure 2c we compare two of the FK506 binding sites and two RAP binding sites. Structures 1tcoC and 1bkf both bind FK506, but while residues Ala 81, His 87 and Ile 91 bind the ligand in the wild type (1tocC) they do not in 1bkf (a double mutant, R42K and H87V). Despite the mutations, ligand flexibility allows the FK506 to bind (27). Structures 1fkb and 1fapA bind RAP at the same binding site with different binding at residue Gln53.
FUTURE DIRECTIONS
Where mapping onto PDB sequences is possible, functional information from different sources may be integrated into FireDB. Candidate sources include Swissprot, PDBsite, dbPTD (Post-translational Modification Database) (28) or PMD (The Protein Mutant Database) (29).
The library of GO terms linked to molecular compounds is being expanded to any ligand where the relation between GO term and chemical compound is clear and this will be added to FireDB in next releases.
Such a big resource of information will be also invaluable for homology based function prediction, we hope to integrate a system that will allow predictions based on automatic transference of conserved binding residues while alignment quality is evaluated.
Acknowledgments
We would like to thank the valuable input and suggestions from David de Juan and Ana M. Rojas also we would like to thank Eduardo Andres and Angel Carro for technical assistance. This work was supported by grants: BioSapiens (LSHG-CT-2003-503265), GeneFun (LSHG-CT-2004-503567), MEC (BIO2004-00875) and EMBRACE (LHSG-CT-2004-512092). Funding to pay the Open Access publication charges for this article was provided by GeneFun (LSHG-CT-2004-503567).
Conflict of interest statement. None declared.
REFERENCES
- 1.Westbrook J., Feng Z., Chen L., Yang H., Berman H.M. The Protein Data Bank and structural genomics. Nucleic Acids Res. 2003;31:489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H.M. The past and the future of structure databases. Curr. Opin. Biotech. 1999;10:76–80. doi: 10.1016/s0958-1669(99)80014-7. [DOI] [PubMed] [Google Scholar]
- 3.Rost B. Marrying structure and genomics. Structure. 1998;6:259–263. doi: 10.1016/s0969-2126(98)00029-x. [DOI] [PubMed] [Google Scholar]
- 4.Pazos F., Sternberg M.J.E. Automated prediction of protein function and detection of functional sites from structure. Proc. Natl Acad. Sci. USA. 2004;101:14754–14759. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd A.E., Orengo C.A., Thornton J.M. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 6.Chandonia J.M., Brenner S.E. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 7.Kleywegt G.J., Jones T.A. Databases in protein crystallography. Acta Cryst. 1998;D54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 8.Laskowski R.A., Chistyakov V.V., Thornton J.M. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acid Res. 2005;33:D266–268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Z., Chen L., Maddula H., Akcan O., Oughtred R., Berman H.M., Westbrook J. Ligand depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20:2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 10.Hendlich M., Bergner A., Gunther J., Klebe G. Relibase: Design and developement of a database for comprehensive analysis of protein-ligands interactions. J. Mol. Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- 11.Stuart A.C., Ilyn V.A., Sali A. LigBase: a databaes of families of aligned ligand binding sites in known protein sequences and structures. Bioinformatics. 2002;18:200–201. doi: 10.1093/bioinformatics/18.1.200. [DOI] [PubMed] [Google Scholar]
- 12.Puvanendrampillai D., Mitchell J.B. L/D Protein Ligand Database (PLD): additional understanding of the nature and specificity of protein–ligand complexes. Bioinformatics. 2003;19:1856–1857. doi: 10.1093/bioinformatics/btg243. [DOI] [PubMed] [Google Scholar]
- 13.Shin,J-M. and Cho,D-H. PDB-Ligand: a ligand database based on PDB for the automated and customized classification of ligand-binding structures. Nucleic Acids Res. 2005;33:D238–D241. doi: 10.1093/nar/gki059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanisenko V.A., Pintus S.S., Grigorovich D.A., Kolchanov N.A. PDBSite: a database of the 3D structure of protein functional sites. Nucleic Acids Res. 2005;33:D183–D187. doi: 10.1093/nar/gki105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter C.T., Bartlett G.J., Thornton J.M. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrance J.W., Bartlett G.J., Porter C.T., Thornton J.M. Using a library of structural templates to recognise catalytic sites and explore their evolution in homologous families. J. Mol. Biol. 2005;347:565–581. doi: 10.1016/j.jmb.2005.01.044. [DOI] [PubMed] [Google Scholar]
- 17.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 18.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 19.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tress M.L., Jones D.T., Valencia A. Predicting reliable regions in protein alignments from sequence profiles. J. Mol. Biol. 2003;330:705–718. doi: 10.1016/s0022-2836(03)00622-3. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velankar S., McNeil P., Mittard-Runte V., Suarez A., Barrel D,, Apweiler R., Henrik K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.IUPAC-IUBMB Joint Commission on Biochemical Nomenclature (JCBN) and Nomenclature Committee of IUBMB (NC-IUBMB). Eur. J. Biochem. 1999;264:607–609. [PubMed] [Google Scholar]
- 24.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponomarnko J.V., Bourne P.E., Shindyalov I.N. Assigning new GO annotations to protein data bank sequences by combining structure and sequence homology. Proteins. 2005;58:855–865. doi: 10.1002/prot.20355. [DOI] [PubMed] [Google Scholar]
- 26.Bourne P.E., Berman H.M., McMahon B., Watenpaugh K.D., Westbrook J., Fitzgerald P.M.D. The macromolecular crystallographic information file (mmCIF) Meth. Enzymol. 1997;277:571–590. doi: 10.1016/s0076-6879(97)77032-0. [DOI] [PubMed] [Google Scholar]
- 27.Futer O., DeCenzo M.T., Aldape R.A., Livingston D.J. FK506 binding protein mutational analysis. Defining the surface residue contributions to stability of the calcineurin co-complex. J. Biol. Chem. 1995;270:18935–18940. doi: 10.1074/jbc.270.32.18935. [DOI] [PubMed] [Google Scholar]
- 28.Lee T-Y., Huang H-D., Hung J-H., Huang H-Y., Yang Y-S., Wang T-H. dbPTM: an information repository of protein post-translational modification. Nucleic Acid Res. 2006;34:D622–D627. doi: 10.1093/nar/gkj083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawabata T., Ota M., Nishikawa K. The Protein Mutant Database. Nucleic Acid Res. 1999;27:355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]