Summary
Ribonuclease P (RNase P) is the ribonucleoprotein endonuclease that processes the 5′-ends of precursor-tRNAs. Bacterial and eukaryal RNase P RNAs had the same primordial ancestor, however, they were molded differently by evolution. RNase P RNAs of eukaryotes, in contrast to bacterial RNAs, are not catalytically active in vitro without proteins. By comparing the bacterial and eukaryal RNAs we can begin to understand the transitions made between the RNA and protein-dominated worlds. We report based on crosslinking studies that eukaryal RNAs, although catalytically inactive alone, fold into functional forms and specifically bind tRNA even in the absence of proteins. Based on the crosslinking results and crystal structures of bacterial RNAs we develop a tertiary structure model of the eukaryal RNase P RNA. The eukaryal RNA contains a core structure similar to the bacterial RNA, but lacks specific features, that in bacterial RNAs contributes to catalysis and global stability of tertiary structure.
Introduction
Ribonuclease P (RNase P) removes 5′-leader sequences from precursor-tRNAs and occurs in all three phylogenetic domains: Bacteria, Archaea and Eukarya. The enzyme is universally composed of both protein and RNA moieties. The RNase P RNAs are all homologs, of common ancestry, but they have undergone substantial evolutionary diversification. The RNA moiety from bacterial and some archaeal holoenzymes can process precursor-tRNA in vitro in the absence of proteins. In contrast, catalysis by the eukaryal RNase P requires multiple protein component(s) in addition to the RNA component. Insights into how the evolutionary process transformed a primordial ribozyme into a ribonucleoprotein can be gained by comparing the bacterial and eukaryal RNA components of RNase P.
The eukaryal RNase P holoenzyme differs from the bacterial and archaeal versions in both protein composition and RNA structure. In vivo, the bacterial RNase P RNA is associated with a single protein. The archaeal and eukaryal holoenzymes are more complex and contain at least four and nine proteins, respectively (Chamberlain et al., 1998; Hall and Brown, 2002; Jarrous, 2002; Kouzuma et al., 2003). In the case of human RNase P, two proteins in addition to the RNA are sufficient for activity in vitro (Mann et al., 2003). It is not known how these proteins contribute to RNase P activity.
Secondary structure models for the eukaryal RNase P RNA have been increasingly well developed through phylogenetic comparisons of diverse eukaryotic RNAs and a core structure shared with bacterial RNAs is evident (Chen and Pace, 1997; Frank et al., 2000; Marquez et al., 2005; Pitulle et al., 1998; Tranguch and Engelke, 1993). The eukaryal RNA seems to be a more rudimentary version of the bacterial RNase P RNA. The typical eukaryotic RNase P RNA is roughly two-thirds the sequence length of the typical bacterial RNA. Although sequence conservation among the eukaryal RNAs is low, it includes distinct sequence elements, Conserved Regions (CRs) I-V, found in all archaea and bacterial RNAs.
Eukaryal RNase P RNAs clearly are homologs of the bacterial RNAs, and consequently are expected to have function and structure in common with those other RNase P RNAs. However, investigation of eukaryal RNase P RNA function in vitro has been limited by the lack of catalytic activity of the RNA alone. In this study, we used photoaffinity crosslinking to investigate the function of the eukaryal RNase P RNA and its global structure. Photoaffinity crosslinking has previously been used to map the active site of bacterial RNase P RNA and to generate distance constraints for inference of the RNA tertiary structure (Burgin and Pace, 1990; Chen et al., 1998; Harris et al., 1994; Nolan and Pace, 1996). The results reported here indicate that several eukaryal RNase P RNAs are capable of binding tRNA specifically and independently of protein, albeit with low affinity. This suggests that the eukaryal RNAs intrinsically possess information sufficient to fold into a structure similar to that of the bacterial RNAs independently of protein. We verify this with intramolecular crosslinking experiments. Data from crosslinking results and the recent bacterial RNA crystal structure were used to infer a specific tertiary structure model for the eukaryotic RNase P RNA. The model indicates that the eukaryal RNase P RNAs lack key elements in the active site and lack stabilizing helices important for proper structure.
Results
5′-and 3′-arylazido-mature tRNA crosslinks to eukaryal RNase P RNA
Photoaffinity crosslinking was used to assay eukaryal RNase P RNAs for the capacity to bind tRNA. The photoagent was attached to the 5′-phosphate of mature tRNA because that is the phosphate acted upon by RNase P and so it is expected to be delivered to the active site of the RNase P RNA. The bacterial RNase P RNAs bind mature tRNA almost as tightly as precursor tRNAs (Reich et al., 1988), so we reasoned by homology that the eukaryal RNA would do so as well, if capable of specific binding. 32P-labeled mature Bacillus subtilis tRNAasp was synthesized by transcription in the presence of guanosine 5′-monophosphorothioate (GMPS), which is incorporated only in the first nucleotide position of the tRNA. An arylazide photoaffinity agent (azidophenacyl bromide) then was coupled to the unique 5′-phosphorothioate of the tRNA. The modified tRNA was incubated with RNase P RNAs from a broad diversity of eukaryotic species and UV irradiated to activate crosslinking. RNase P RNAs were in vitro transcripts from the corresponding genes from Homo sapiens, Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, Schizosaccharomyces pombe and Acanthamoeba castellanii. Crosslinked products were separated from unreacted substrate by polyacrylamide gel electrophoresis as shown in Figure 1A.
Figure 1.
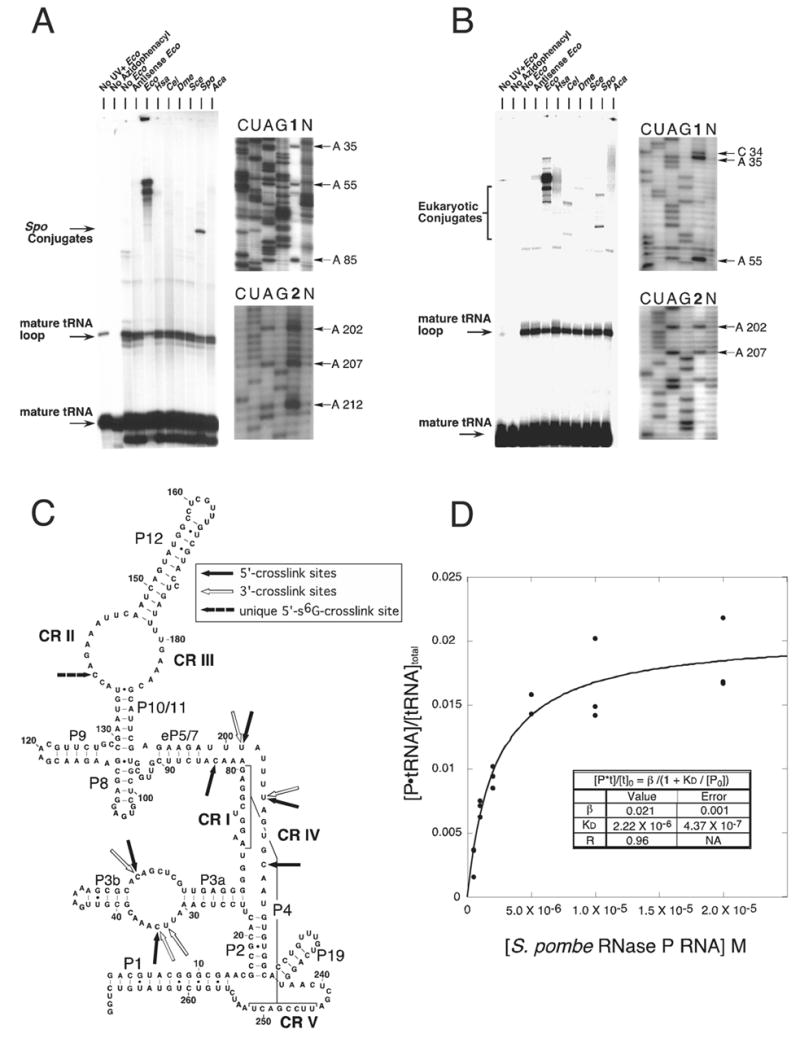
Intermolecular crosslinking analysis of eukaryal RNase P RNA-tRNA conjugates. (A) 5′-arylazido-B. subtilis mature tRNAasp crosslinks to eukaryal RNase P RNA. All reactions are identical except: (1) The reaction contained E. coli RNase P RNA but was not exposed to UV, (2) the thio-containing tRNA was not coupled to the azidophenacyl bromide, (3) no E. coli RNase P RNA, (4) RNA complementary to E. coli RNase P RNA was included instead of E. coli RNase P RNA, (5) E. coli RNase P RNA, (6) H. sapiens RNase P RNA, (7) C. elegans RNase P RNA, (8) D. melanogaster RNase P RNA, (9) S. cerevisiae RNase P RNA, (10) S. pombe RNase P RNA and (11) A. castellanii RNase P RNA was included. The S. pombe RNase P RNA-5′-arylazido-tRNA conjugates were analyzed by primer extension. Unlabeled S. pombe RNase P RNA-tRNA conjugates were prepared, purified and quantified as described in Experimental Procedures. Lane 1 and 2 contain primer extension products using oligonucleotides 150R and 250R respectively. Lanes C, U, A, and G correspond to sequencing reactions with non-crosslinked RNA template, lane N is a control primer extension without dideoxynucleotides of unmodified S. pombe RNase P RNA. The termination sites of primer extension are indicated to the right of each gel. (B) 3′-arylazido-mature RNA crosslinks to eukaryal RNase P RNA. Crosslinking and gel analysis is identical to A. (C) The secondary structure of S. pombe RNase P RNA with the crosslink sites inferred from primer extension analysis indicated by arrows. An RNase P RNA structural nomenclature is described in Marquez et al. 2005. Solid arrows indicate sites in the S. pombe RNA that crosslink to the 5′-end of tRNA. Unfilled arrows indicate sites in the S. pombe RNA that crosslink to the 3′-end of tRNA. The dashed arrow indicates the unique site crosslinked by 5′-s6G-labeled tRNA. (D) Measurement of the dissociation constant (Kd) of arylazido-mature B. subtilis tRNAasp and S. pombe RNase P RNA. Crosslinking reactions were performed in the presence of increasing amounts of S. pombe RNase P RNA (0 –2.0 X 10−5 M) incubated with 32P-labeled 5′-arylazido-mature tRNA (228 nM). (•) represents the fraction of tRNA bound to S. pombe RNase P RNA as assayed by radioactivity in the crosslinked band. The data were fit to the binding isotherm equation in Experimental Procedures.
In general, more slowly migrating bands correspond to RNase P RNA-tRNA conjugates. Among those RNAs tested for binding tRNA, crosslinked products were observed only with the S. pombe RNA and the Escherichia coli bacterial positive control. The extent of crosslinking in the case of the E. coli RNA was more than 10-fold greater than with the S. pombe RNA. It is possible that the other eukaryal RNAs do bind tRNA, but irradiation does not give rise to crosslinked products (see below). To verify that crosslinking occurred through the photoaffinity agent and not between random parts of the RNAs due to UV exposure alone, we used phenylmercuric acetate to dissociate the E. coli and S. pombe RNase P RNA-tRNA conjugates (Burgin and Pace, 1990); organomercurials specifically cleave sulfur linkages (Neumann et al., 1967). Upon incubation with phenylmercuric acetate, all conjugates dissociated into RNase P RNA and tRNA (data not shown). Thus, the crosslinked products were due to the arylazide group linked to the 5′-end of tRNA via sulfur.
We used the crosslinking assay to test some additional aspects of the binding reaction. We determined that precursor-tRNA crosslinks to S. pombe RNase P RNA with approximately the same efficiency as mature-tRNA (Supplemental Figure 1SA), indicating that the 5′-leader sequence does not contribute significantly to the binding of tRNA. In order to determine if S. pombe RNase P RNA discriminates between eukaryal and bacterial tRNAs, a photoaffinity agent was placed at the 5′-end of S. cerevisiae 32P-labeled mature-tRNAphe and then crosslinked to S. pombe RNase P RNA. S. cerevisiae tRNAphe crosslinked to S. pombe RNase P RNA with the same efficiency as B. subtilis mature-tRNAasp (Supplemental Figure 1SB). These data indicate that eukaryotic RNase P does not discriminate between bacterial and eukaryal tRNAs.
The eukaryotic RNase P RNAs require cations to bind tRNA. When monovalent and divalent salts were omitted from the crosslinking reactions, no crosslink products were detected (data not shown). A titration of increasing monovalent salt (NH4+), 0 to 5.0 M, with the divalent salt (Mg2+) concentration constant at 25 mM, gave identical crosslink products and yield for each reaction. A titration of increasing divalent salt (Mg2+), 0 to 1.0 M, with the monovalent salt (NH4+) concentration constant at 3.0 M, also gave identical crosslink products and yield for each reaction (data not shown). Thus, in the presence of minimum cation concentrations, binding is fairly insensitive to ionic strength. Binding of the tRNA by S. pombe RNase P RNA does not depend on Mg2+, as is also the case with bacterial RNase P RNA (Beebe et al., 1996; Smith et al., 1992).
In addition to the arylazide group, which is 9 Å in length, we employed in crosslinking experiments another, shorter photoaffinity agent, 6-thioguanosine (s6G) (1.5 Å) (Favre et al., 1998). This photoagent was incorporated during transcription, site-specifically at the 5′-end of the tRNA. The resulting tRNA was submitted to crosslinking analysis with the same RNase P RNAs as above. As with the 5′-azido-labeled tRNA, crosslinked products were observed only with the E. coli and S. pombe RNAs (Supplemental Figure 2S).
We also analyzed the ability of the various eukaryotic RNase P RNAs to bind tRNA modified to contain an arylazide photoaffinity agent (4-azidobenzoyl) at the 3′-terminus (Oh and Pace, 1994). As a control, parallel crosslinking experiments were carried out with E. coli RNase P RNA, which resulted in products identical to those previously reported (Oh and Pace, 1994). No conjugates occurred in the absence of RNase P RNA or photoaffinity agent. Although the extent of crosslinking to the eukaryotic RNAs was very low, apparent conjugate bands nonetheless occurred with the H. sapiens, C. elegans, D. melanogaster, and S. pombe RNAs, but not with the S. cerevisiae RNA (Fig. 1B). The reaction with the A. castellanii RNA did not yield defined bands for unknown reasons. The observation that 3′-photoagent-labeled tRNA formed crosslinks with most eukaryal RNase P RNAs tested, whereas the 5′-photoagent-tRNA does not, suggests that the positioning of the 5′-end of tRNA in these cases was unstable or too distant from the RNase P RNAs to crosslink. The only 3′-conjugate with sufficient material for use in mapping crosslinked sites was that of S. pombe.
To identify the tRNA crosslink sites in the S. pombe RNase P RNA, primer extension analyses were conducted on conjugates eluted from gels. The results of these analyses with 5′-and 3′-arylazido-tRNAs are shown in Figure 1. 5′-s6G-tRNAs analysis are shown in Supplemental Figure 2S. Dideoxynucleotide-terminated RNA sequencing lanes provide a register for the primer extension products. Primer extension on unmodified S. pombe RNA revealed nucleotides where reverse transcriptase paused or terminated spontaneously, presumably due to structure in the RNA template (Lane et al., 1985). Pauses not seen in the unmodified S. pombe RNase P RNA are true crosslinks. Because reverse transcriptase cannot traverse the crosslink site, the last nucleotide incorporated is interpreted as one nucleotide prior to the actual site of the crosslink (Ehresmann et al., 1987).
All the crosslinking data for S. pombe RNase P RNA are summarized in Figure 1C. Crosslinks occurred in CR II, IV and in regions of the eukaryotic RNA that are not conserved in the bacterial RNAs. The crosslinking into these CRs suggests the functional importance of the interaction (Burgin and Pace, 1990; Harris and Pace, 1995; Kazantsev et al., 2005; Pagan-Ramos et al., 1996). Primer extension analysis of the only resolvable conjugate formed between the S. pombe RNase P RNA, and 5′-s6G-tRNA indicated that three of the four premature terminations (A202, A207, and C212) were identical to those found when 5′-azidophenacyl tRNA was used for crosslinking. The unique termination site was at A139. The reason the 5′ and 3′ crosslink sites are in close proximity to each other likely is because the 5′ and 3′ ends of tRNA are adjacent to one another in the tRNA acceptor stem. These results indicate that eukaryal RNase P RNAs, although catalytically inactive, specifically bind tRNA in the absence of proteins. The crosslink sites in the eukaryal RNA are similar to those seen with the bacterial RNA, near the eukaryotic homolog of the bacterial active site (see below).
Binding affinity of S. pombe RNase P RNA for mature tRNA
We used the crosslinking assay to evaluate the dissociation constant for the complex between the S. pombe RNase P RNA and 5′-arylazido-mature tRNA. A constant concentration of 32P-labeled 5′-arylazido-tRNA was incubated with different concentrations of S. pombe unlabeled RNase P RNA and then irradiated with UV. The resulting complexes were resolved by denaturing polyacrylamide gel electrophoresis as above. We assume that crosslinking occurs at equilibrium, so the crosslinked conjugate is expected to be proportional to the concentration of the RNase P RNA-tRNA complex at the time of UV exposure. Crosslinking reactions were conducted in triplicate at each RNA concentration and the data were in good agreement with the binding isotherm equation shown in Figure 1D. We extract from the data a dissociation constant (Kd) of ~ 2.2 μM. The binding is specific: nonlabeled mature tRNA competes with 5′-arylazido-tRNA, while unlabeled antisense E. coli RNase P RNA is unable to compete (data not shown). This means that the eukaryal RNase P RNA is capable of part of the catalytic cycle, binding of substrate.
Eukaryotic RNase P RNAs are Structurally Less Stable than Bacterial RNAs
The eukaryotic RNase P RNAs typically lack hairpin elements that in bacteria are known to contribute to the stability of the tertiary structural folding of the RNA. This suggests that the global packing of the eukaryotic RNAs might be less stable than that of bacterial RNase P RNAs, perhaps the cause of the inactivity of the eukaryotic RNAs. To examine the conformations of the eukaryotic RNase P RNA compared to the bacterial RNAs, we used native polyacrylamide gel electrophoresis. In the conditions of 100 mM NH4+ and 0, 0.5, 1.0 and 5.0 mM Mg2+ all RNAs migrated mainly as single bands (Fig. 2A), indicating hydrodynamic and, thereby, conformational homogeneity under the particular conditions. However, the eukaryotic and bacterial RNAs respond differently to elevated Mg2+, as summarized in Figure 2A. The bacterial RNAs both display an increase in relative mobility, an apparent compaction, between 0 and 5.0 mM Mg2+. This would be consistent with a tightening of tertiary structure as Mg2+ ions counteract electrostatic repulsion due to the phosphodiester chain. In contrast, none of the eukaryal RNAs undergoes appreciable change in electrophoretic mobility at higher Mg2+ concentration (Fig. 2A). This would be consistent with the notion that the eukaryal RNAs cannot adopt the stable tertiary structure equivalent to their bacterial homologs and so are not amenable to further compaction. Thus, the Mg2+-independent electrophoretic properties of the eukaryal RNAs may reflect lack of assembly of the core helices to form the tertiary structure that in the bacterial RNAs is stabilized by hairpin elements not present in the eukaryal RNAs.
Figure 2.
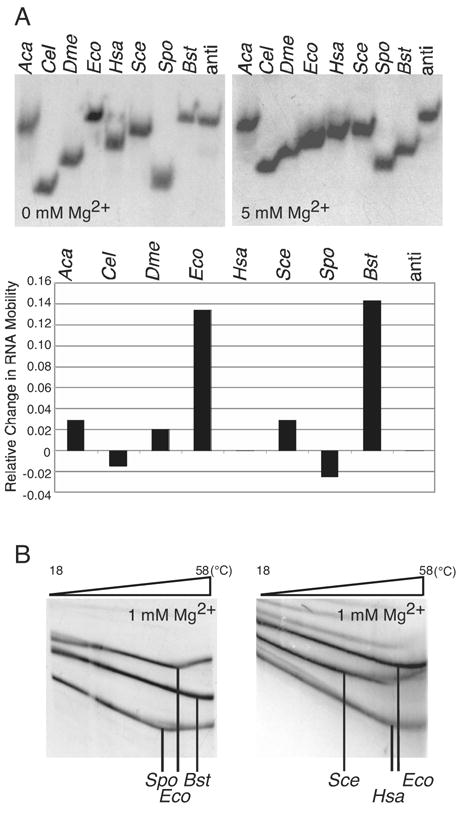
(A) Eukaryotic RNase P RNAs are affected differently by Mg2+ and are less stable than bacterial RNase P RNAs. Various eukaryotic and bacterial RNase P RNAs (Aca, A. castellanii; Cel, C. elegans; Dme, D. melanogaster; Eco, E. coli; Hsa, H. sapiens; Sce, S. cerevisiae; Spo, S. pombe; Bst, B. stearothermophilus; anti, control antisense E. coli) were folded and analyzed on 4.5% acrylamide 1X THE native gels containing 100 mM NH4OAc and various concentrations of Mg2+ (0 and 5 mM, shown left and right respectively). Minor amounts of some eukaryotic RNase P RNAs migrated in oligomeric forms as seen with bacterial RNAs and previously reported in Buck et al., 2005b (not shown). The relative change in RNA mobility between the 0 mM and 5 mM Mg2+ gels, normalized to the anti Eco control RNA, is graphed below. Relative change in mobility is defined as: (d5mM/anti d5mM)/(d0mM/anti d0mM) – 1, where d5mM is the distance the RNA traveled from the well on the 5 mM Mg2+ gel, anti d5mM is the distance the anti Eco control RNA traveled from the well on the 5 mM Mg2+ gel, d0mM is the distance the RNA traveled from the well on the 0 mM Mg2+ gel, and anti d0mM is the distance the anti Eco control RNA traveled from the well on the 0 mM Mg2+ gel. (B) The thermal stability of various eukaryotic and bacterial RNase P RNAs was analyzed by temperature gradient gel electrophoresis (TGGE). Gel conditions are as in A, except containing 1 mM Mg2+. (left) The E. coli and B. stearothermophilus RNase P RNAs melt at a higher temperature than S. pombe RNase P RNA. (right) Similarly, the E. coli RNase P RNA melts at a higher temperature than either H. sapiens or S. cerevisiae RNase P RNA. Vertical lines indicate the transition temperature, where RNA tertiary structure unfolds.
We additionally tested the relative thermal stabilities of the eukaryotic RNase P RNAs compared to their bacterial homologs using temperature gradient gel electrophoresis (TGGE). With this method RNAs electrophoretically migrate perpendicular to a linear temperature gradient. When RNA structure changes from a compact to a more extended, denatured conformation, electrophoretic mobility slows. As shown in Figure 2B, 32P-labeled Bacillus stearothermophilus, E. coli and S. pombe RNase P RNAs were subjected to TGGE from 18 to 58°C. The S. pombe RNase P RNA begins to unfold at 45 °C, while the bacterial RNAs shift at higher temperatures. Given that secondary structures nominally unfold at temperatures greater than 60 °C (Buck et al., 2005), the unfolding detected in this temperature range is expected to reflect change in tertiary structure. Similar analysis was conducted with all eukaryotic RNAs used in this study, with similar results (Figure 2B and Supplemental Figure 3S). Thus, the tertiary structures of the eukaryal RNase P RNAs are systematically less stable to thermal denaturation than are the bacterial RNAs.
Intramolecular photoaffinity crosslinking analysis
The ability of the S. pombe RNase P RNA to bind tRNA, albeit weakly, implies that the structure of the RNA is ordered when complexed with substrate, even though it is catalytically inactive. To test if the S. pombe RNase P RNA adopts a specific tertiary structure in the absence of substrate, intramolecular crosslinking experiments were conducted and the results were compared to previous crosslinking studies with bacterial RNase P RNAs. To generate new 5′-and 3′-ends at specific sites within the sequence of the S. pombe RNase P RNA, we used circularly permuted RNAs (cpRNAs) (Thomas et al., 2000). The azidophenacyl crosslinking agent was attached to the 5′-end of the cpRNA. Six circularly permuted S. pombe RNase P RNAs, cp60, cp69, cp118, cp140, cp210, and cp242, (e.g. cp60 signifies the 5′-end of cp60 begins at native nucleotide 60) were made for site-specific crosslinking analysis. These photoagent-attachment sites in the S. pombe cpRNAs are homologous to those previously subjected to similar analysis with the bacterial RNAs. The introduction of internal nicks in RNAs usually has no impact on the folded structures (Kazantsev et al., 2005; Pan et al., 1991). In our study the cpRNAs migrated as single bands in native gels (100 mM NH4+ and 1.0 mM Mg2+) and retained the capacity to bind tRNA in the crosslinking assay (Supplemental Figure 4S). cpRNAs therefore adopt the functional structure.
Intramolecular crosslinking experiments were conducted under conditions determined to be optimal for intermolecular crosslinking reactions (above). Two classes of crosslinks form in the intramolecular crosslinking reaction; circular and lariat molecules (Harris et al., 1997). Circular molecules, usually the most slowly migrating bands in gels, result from crosslinking from the photoagent to the nearby 3′-end of the same RNA molecule and so provide little useful structural information. Lariat molecules, however, result from crosslinking to sites interior to the same RNA molecule. Mapping of these crosslink sites provides distance constraints with which to orient the structural elements of the S. pombe RNA and thereby to compare the tertiary structure of the S. pombe RNA to the previously determined bacterial structure.
UV irradiation of the photoagent-containing, circularly permuted S. pombe RNase P RNAs generally yielded strong crosslinks (Fig. 3A). The occurrence of discrete crosslinks implies that the S. pombe RNA adopts a specifically folded structure even in the absence of any protein component. Crosslinked RNAs were excised from preparative gels and the locations of crosslinks were identified by primer extension as illustrated in Figure 3B. The crosslinking patterns of photoagents placed within the conserved core of the S. pombe RNase P RNA generally are similar to those obtained with bacterial RNAs. For instance, intramolecular crosslinking results with cp69 and cp242 RNAs demonstrate the close proximity of CR I, CR IV and CR V in the S. pombe RNA, and correspond to crosslinks seen with bacterial RNase P RNA. As in the E. coli and B. subtilis RNAs, the loop of P9 in the S. pombe RNA is in close proximity to P1 (Chen et al., 1998). These corroborative data between the bacterial and the eukaryal RNAs indicate that eukaryal RNase P RNA forms a self-folded core structure similar to that of the bacterial ribozyme.
Figure 3.
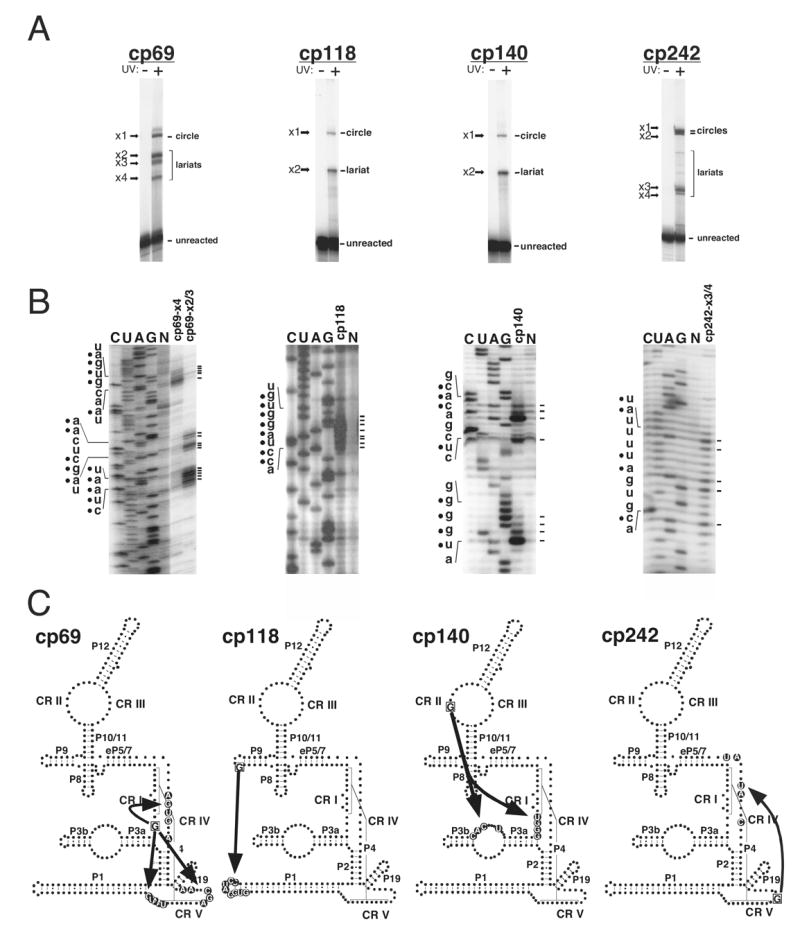
Intramolecular crosslinking analysis of circularly permuted S. pombe RNase P RNAs, cp69, cp118, cp140 and cp242. (A) Uniformly 32P–labeled photoagent-containing cpRNAs without (UV-) or with (UV+) 302 nm UV irradiation were analyzed by gel electrophoresis and autoradiography. The crosslinking reactions contained 1.0 μM photoagent containing RNAs. The cpRNA crosslink reactions resulted in crosslinked bands, designated x1, -x2, -x3 etc. (B) Primer extension mapping of crosslinked RNAs, were carried out with 5′-32P-labeled oligonucleotides complementary to S. pombe RNase P RNA (Supplemental Experimental Procedures). Primer extension mapping of some crosslinked RNAs indicated that they were circular molecules and uninformative (data not shown). Lanes C, U, A, and G correspond to sequencing reactions with non-crosslinked RNA template, lane N is a control primer extension without dideoxynucleotides of unmodified S. pombe RNase P RNA. The primer extension reactions using the crosslinked species are indicated above each lane. The termination sites of primer extension are indicated to the right of the gel. The RNA sequence of the termination sites is shown on the left. Filled circles denote the actual crosslink sites which are one nucleotide 5′ to the primer extension termination sites. (C) Crosslinking sites in the RNA secondary structures are shown. The boxed G indicates the photoagent attachment site located at the 5′ end of the cpRNA. The circled bases in the secondary structure represent the corresponding crosslinking sites. Arrowheads indicate the direction of the crosslinks.
A few results were not useful or differed between the bacterial and eukaryal RNAs. For instance, crosslinking of cp60 resulted only in local crosslinks, within the P3 bulge loop, providing no useful structural information. Unexpectedly based on the bacterial structure, when the photoagent is located in CR II (cp140) it forms crosslinks to nucleotides in CR I and to nucleotides in the internal loop of P3 (Figure 3C). These results would indicate that CR II and the internal loop of P3 are in close proximity to CR I. However, compared to the crystal structures of Thermotoga maritima and B. stearothermophilus RNase P RNAs, CR II and CR I are not within crosslinking distance of each other (see Discussion) (Kazantsev et al., 2005; Torres-Larios et al., 2005). Nonetheless, the similarities of most of the bacterial and eukaryal crosslinking results, and the clear homology of the RNAs justify specification of a working model for the tertiary structure of the S. pombe RNase P RNA based on the crystal structure of the bacterial RNA from B. stearothermophilus (Kazantsev et al., 2005).
Computer modeling of S. pombe RNase P RNA
A model of the tertiary structure of the S. pombe RNA was inferred by homologous replacement and is shown in comparison to the bacterial structure in Figure 4. 195 nucleotides out of 271 in the S. pombe RNA could be reliably modeled from the alignment of the B. stearothermophilus and S. pombe RNase P RNA sequences. The accuracy of the homologous modeling approach has been confirmed by the correspondence of comparative and crystal structures for 16S and 23S rRNAs (Gutell et al., 2002). Comparison of the bacterial and eukaryal RNase P RNA structures shows that both share a similar core structure, but the eukaryal RNA lacks peripheral helices known to stabilize the bacterial RNA structure (e.g. B. stearothermophilus P5.1-P15.1; E. coli P14-P8), as well as nucleotides known to be important for binding and catalysis (e.g. B. stearothermophilus and E. coli. L15) (Brown et al., 1996; Darr et al., 1992; Kirsebom and Svard, 1994). In addition, the internal loop of P3, missing in bacterial RNAs, lies near the bacterial P15, which does not occur in the eukaryotic RNAs. Of the 45 nucleotides that comprise the B. stearothermophilus RNase P RNA catalytic core, shown in Figure 5, at least 37 homologous nucleotides are found in the S. pombe RNA. Of the 13 base pairs identified in the B. stearothermophilus core, all are present in the S. pombe RNA as well. Although the sequences in these base pairs vary, compensatory base substitutions found in the S. pombe RNA should not perturb the tertiary structure in the bacterial model because they are isosteric with the bacterial base pairs (Leontis et al., 2002). The only possible variation to isostericity rules is S. pombe A249:G74 which is a C:G pair in B. stearothermophilus (Figure 5). However, this P4 base pair also is likely isosteric, because P4 is a highly conserved helix and the homologous base pair in other eukarya indeed are canonical Watson-Crick base pairs. The structural relationships of three nucleotides in J4/1 of the S. pombe RNA (A253, U254 and C255) are uncertain and therefore are not paired in the model. However, because of positional homology with the comparable bacterial nucleotides, they likely form the same structure seen in the bacterial RNA.
Figure 4.
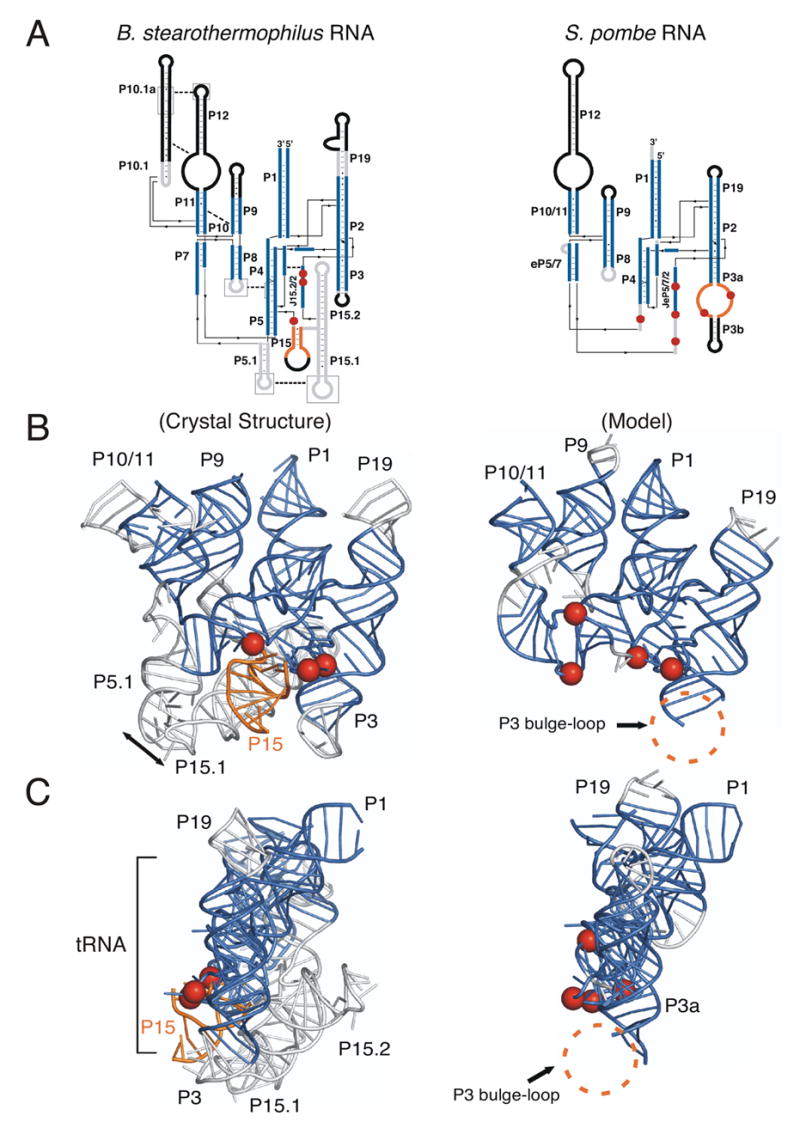
Commonalities in structure and function between the crystal structure of bacterial RNase P RNA and the modeled eukaryal RNase P RNA. (A) Coaxial stack representation of the secondary structures of B. stearothermophilus and S. pombe RNase P RNAs. The RNA that is homologous between B. stearothermophilus and S. pombe RNase P RNAs are represented as blue, non-homologous RNA is colored gray and RNA not represented in the B. stearothermophilus crystal structure is colored black in both molecules. Arrows indicate the 5′ to 3′ direction. Sites of 5′-tRNA crosslinking are represented as red spheres. B. stearothermophilus long-range tertiary interactions between helices are indicated by dashed lines, while homologous interactions are lacking in S. pombe RNA. The main site of 3′-tRNA crosslinking (P15) in B. stearothermophilus RNA is colored gold. A main site of 3′-tRNA crosslinking in S. pombe RNA is the P3 bulge-loop and, while not homologous to bacterial P15, is also colored gold. (B) Tertiary structure ribbon models of B. stearothermophilus and S. pombe RNase P RNAs, as colored in A. S. pombe RNase P RNA nucleotides 136-185 are not included in the modeling because of the inconsistency of the results between cp140 crosslink sites and the crystal structure of T. maritima RNase P RNA. The double-headed arrow indicates the long-range tertiary interaction between P5.1 and P15.1. The structure of P3 bulge-loop and P3b could not be reliably inferred from the bacterial structure and therefore has not been modeled. However, the location of 5′ and 3′-tRNA crosslinks (indicated by red spheres and regions highlighted in gold) suggest that these regions of the S. pombe RNA are in close proximity, consistent with the model. (C) Side-view of B. stearothermophilus and S. pombe RNase P RNAs. In the side-view the coaxially stack of P19, P2 and P3 is in the forefront. The general position of tRNA is indicated by a bracket.
Figure 5.
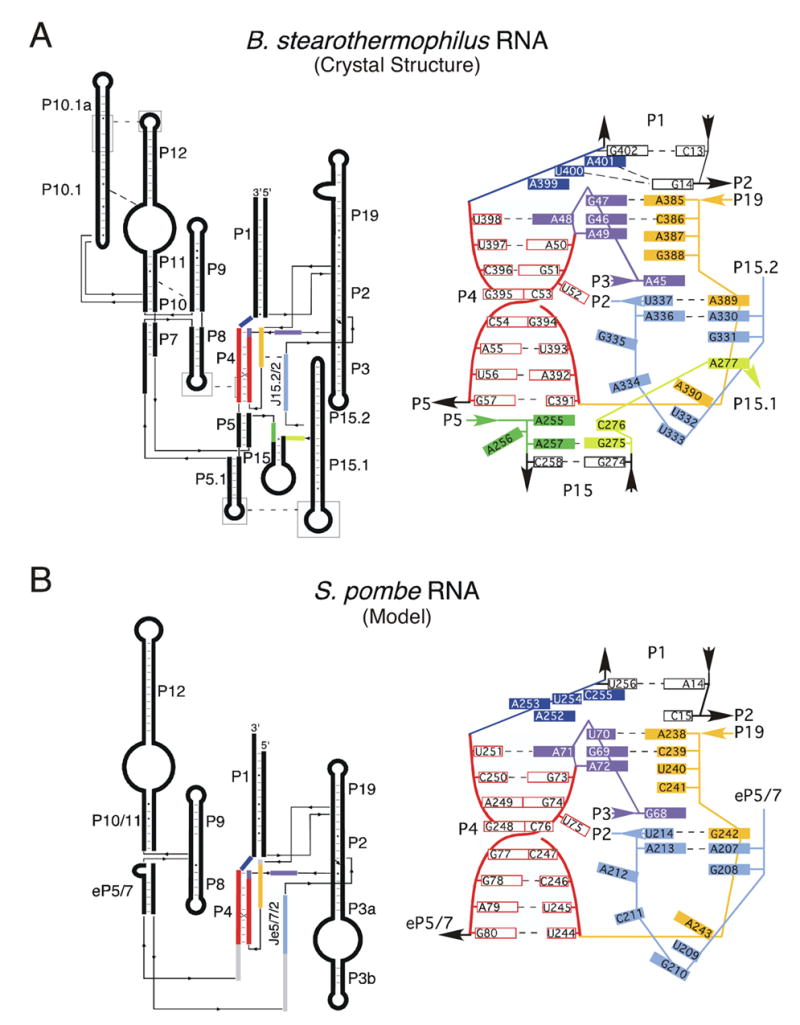
Catalytic core comparison of the bacterial RNA crystal structure and the modeled eukaryal RNase P RNAs. (A) Coaxial stack secondary structure of B. stearothermophilus RNase P RNA and a slab diagram representing base pairing and stacking interactions in the catalytic core. Joining region between P3 and P4 (J3/4) is colored in purple, P4 in red and purple, J5/15 in green, J15/15.1 in yellow, J15.2/2 in light blue, J19/4 in orange and J4/1 in dark blue. (B) Coaxial stack secondary structure of S. pombe RNase P RNA and a slab diagram representing base pairing and stacking interactions in the catalytic core. The NIHMS has received the file ‘mmc1.pdf’ as supplementary data. The file will not appear in this PDF Receipt, but it will be linked to the web version of your manuscript.
Discussion
In this study we used photoaffinity crosslinking to investigate the structure and function of eukaryotic RNase P RNA. We show that the eukaryal RNAs, although catalytically inactive under conditions in which bacterial RNase P RNAs are active, are capable of specifically binding tRNA. We additionally show that the S. pombe RNase P RNA adopts a near-native conformation in the absence of proteins. Utilizing this information as well as the recently solved bacterial RNase P RNA structure we have modeled the structure for a eukaryal RNase P RNA. The model allowed us to see, in detail not previously possible, some likely reasons for the inactivity of the eukaryal RNA alone and suggests possible roles for the required protein subunits.
The S. pombe RNase P RNA structure model (Figure 4) can be interpreted functionally by homology with the bacterial structure. In bacterial as well as eukaryal RNase P RNAs, structural elements thought to be involved in catalysis include P4, CR I, CR IV and CR V. These elements constitute the phylogenetically conserved structural core and face the substrate. In bacterial and archaeal RNase P RNAs, hairpin structures are attached to the phylogenetically conserved core and dock elsewhere to form struts that stabilize the global conformation of the RNA (Pace and Brown, 1995). An example of such an interaction is between P5.1 and P15.1 in Bacillus-type RNAs, shown in Figure 4. Comparable stabilizing structures are absent from the eukaryal RNAs. Some non-conserved hairpin elements occur in some eukaryal RNase P RNAs, however, comparative studies indicate that these are not homologs of the stabilizing helices found in bacterial RNAs (Marquez et al., 2005). It is evident from the crystal structure of the bacterial RNA, and the eukaryal model derived from it, that structural elements attached to the core are positioned to expand in length without distortion of the active core elements. Presumably, in eukaryal holoenzymes one role of the multiple protein components is to assume structural functions that in bacterial holoenzymes are performed by RNA structural elements, for instance to stabilize the global structure.
We used native gel electrophoresis to investigate the effects of Mg2+ and temperature on the folding and stability of the eukaryotic RNase P RNA structures. As Mg2+ concentration is increased, eukaryotic RNAs undergo little change in electrophoretic mobility, whereas the bacterial RNAs undergo an increase in mobility consistent with further compaction of the global structures. Since Mg2+ generally stabilizes RNA structure we interpret the influence of Mg2+ on the mobility of the bacterial RNAs compared to the eukaryal RNAs as a manifestation of structural ordering that the bacterial but not the eukaryal RNA can undergo independently of protein. One explanation for this difference in electrophoretic behavior would be the stabilizing influence of long-range interactions on the bacterial RNA tertiary structure and their absence in the eukaryotic version of the RNA. The thermal gradient gel results are consistent with the relative instability of the global tertiary structures of the eukaryal RNAs compared to the bacterial RNAs: the eukaryal RNase P RNA structures denature at lower temperatures than do the bacterial RNAs.
The less stable structures of the eukaryotic RNase P RNAs compared to the bacterial versions may result in increased potential for misfolding of the eukaryotic RNAs, which may explain a few minor disagreements in the crosslink data obtained with the eukaryal and bacterial RNAs. Intermolecular crosslinking results in the S. pombe RNA with 5′-s6G-tRNA, and intramolecular crosslinking results with cp140, both place CR II near the 5′-end of tRNA and near CR I, IV and V. A photoagent in the homologous position in the bacterial RNA, near CR II, crosslinked only locally (unpublished data). Furthermore, in the crystal structures of the bacterial RNase P RNAs CR II is not in proximity to CR I (Kazantsev et al., 2005; Torres-Larios et al., 2005). Additionally, the 5′-end of tRNA crosslinks to S. pombe RNase P RNA in locations that are not supported by comparable bacterial crosslink sites (Figure 4A). It is possible that, without structurally stabilizing hairpins comparable to those in the bacterial RNAs, eukaryal RNAs are to some extent conformationally heterogeneous in the absence of proteins. This again would be consistent with a role for proteins in stabilization of parts of the molecule that in the case of the bacterial RNase P are stabilized by RNA structure.
An enzymatic reaction can be considered in essence as two steps: one, the binding of substrate; and two, chemical action. Clearly the eukaryotic RNase P RNA is capable of binding substrate, however cleavage of the scissile bond does not occur. The clear homology between the eukaryal and bacterial RNase P RNAs is strong evidence that the eukaryotic RNA, like the bacterial RNAs performs an essential role in the catalytic function of the holoenzyme. The structural model can address the failure of the eukaryal RNA to carry out the catalytic step. The S. pombe RNase P RNA contains most of the catalytic core found in bacterial RNAs as seen in Figure 5. Importantly, the only bases missing from the S. pombe RNase P RNA core compared to the bacterial catalytic core are the 3′ flanking sequences (3 nucleotides) of the bacterial helix P15 and nucleotides homologous to A255 and A256 (B. stearothermophilus numbering). In the bacterial RNA, mutants of the homolog of A255 show a significant decrease in binding affinity and catalysis. Additionally, nucleotide A255 in the bacterial RNA has been shown to interact with the base immediately 5′ of the cleavage site of tRNA (Zahler et al., 2003). Mutations of A256 show little effect on catalysis but show defects in RNA structure near the active site. The S. pombe RNase P RNA catalytic core, while containing most of the catalytic core found in bacteria, clearly lacks bases thought to be critical for binding and catalysis in bacterial RNAs.
In addition, the S. pombe and other known eukaryotic RNase P RNAs lack a homolog of the bacterial P15 and L15, the binding site for the 3′-CCA of tRNAs and potentially an important part of the active site in the bacterial RNA (Kirsebom, 2002; Oh and Pace, 1994; Oh et al., 1998). In eukaryal RNAs, the homologous position is empty or occupied by a region that is particularly variable in length and sequence and is not homologous with the bacterial P15/L15. We used the 3′-photoagent-labeled tRNA as a probe to identify residues in the eukaryal RNase P RNA that are in the vicinity of the 3′-end of tRNA. The 3′-end of tRNA crosslinks to the S. pombe RNA near the location of P15 in the bacterial RNA and to a eukaryote-specific structure, the bulge-loop of P3 (Figure 1C). Notably, the 5′-end of tRNA also crosslinks to this bulge-loop. These results suggest that this eukaryote-specific bulge-loop is near the active site of the RNA. In bacterial RNase P RNA the 5′-photoagent in tRNA crosslinks to P15/L15 (Burgin and Pace, 1990). According to the eukaryal tertiary structure model (Figure 4), the bulge-loop in P3 is located near the position of the bacterial helix P15/L15. The absence of a bacterial P15 from the eukaryotic version nominally provides an empty space that could be occupied by the bulge-loop in P3 and possibly a protein component in vivo (Ziehler et al., 2001).
The absence of the bacterial P15/L15, 3′-CCA binding site from the eukaryotic RNase P RNAs may account for some of the lack of affinity for tRNA and it is unknown what role proteins may have in binding the substrate (Jarrous, 2002). In S. pombe, in vivo, the 3′-CCA sequences of the tRNAs are deposited after RNase P RNA cleaves the 5′-end of pre-tRNA (Van Horn et al., 1997). If this order of processing is true for all eukaryotes then the eukaryotic RNase P RNA may not require a structural homolog of the bacterial loop, at least for binding CCA (Dubrovsky et al., 2004).
Eukaryal RNase P RNA homologs are expected to possess commonalities in function and structure with the bacterial RNase P RNAs. These common structures must have been established early in evolution and have persisted in all domains of life, throughout time. Our results show that the eukaryal core contains the requisite information, independently of protein, to determine the near-native fold for the RNase P RNA, and to bind substrate, the first step in catalysis. The eukaryal and bacterial RNase P RNAs had common primordial ancestry, however they followed different evolutionary paths, during the course of which the eukaryotic RNAs became dependent on protein for catalytic function. Our eukaryal tertiary structure model in comparison with the bacterial structure clearly identifies nucleotides and helical elements that in evolution were lost from the eukaryal RNA or never acquired. Instead, evolution of the eukaryal RNase P incorporated multiple proteins to maintain catalytic activity.
Experimental Procedures
Formation of RNase P RNA-mature tRNA Conjugates
In the standard intermolecular crosslinking reaction, RNase P RNAs were folded prior to crosslinking; 100 ng of photoagent-containing mature tRNA (Supplemental Experimental Procedures) and 4μg RNase P RNA in 25μl of 50 mM Tris-HCl, pH 8.0, 100 mM NH4OAc, 0.05% SDS were incubated for 10 min at 65 °C followed by adding MgOAc to 25 mM and incubated at 37°C for 15 min. Crosslinking reactions were performed essentially as described previously (Burgin and Pace, 1990). Crosslinking was done at 2 cm for 30 min. The crosslinking reaction products were ethanol precipitated and resolved on a 4% polyacrylamide-8 M urea gel. For analytical gels, the crosslinked products were detected by autoradiography and phosphorimager. For preparative gels, the crosslinked RNAs were excised from ethidium bromide-stained gels, eluted into elution buffer at 4 C overnight and RNaseethanol P RNA crosslinks to mature tRNA were cleaved with mercury essentially as described (Burgin and Pace, 1990).
Determination of Kd
To measure the dissociation constant, S. pombe RNase P RNA at various concentrations (5.0 X 10−7, 1.0 X 10−6, 2.0 X 10−6, 5.0 X 10−6, 10−5, 2.0 X 10−5 M) and 32P-labeled arylazido-mature tRNA (228 nM) were prepared as described, incubated together 5 min. at 37 C, then placed on ice. The crosslinking reaction was performed as described above for the formation of RNase P RNA-mature tRNA conjugates. RNAs were resolved by electrophoresis through an 8% polyacrylamide-8 M urea gel. Following UV-irradiation each labeled crosslink band was quantified using a phosphorimager. Background was determined by measurement of radioactivity in a lane in the absence of RNase P RNA (cpm background). The fraction of tRNA bound ([P•tRNA]/tRNAtotal) was calculated as (cpm crosslink band - cpm background)/(cpm tRNA total - cpm background), where cpm is radioactivity in counts per minute. We assume that crosslinking is much faster than the rate of equilibrium, so at equilibrium [P•tRNA] = [crosslink band], and the complex reacts to form a crosslink with efficiency β, 0< β≤ 1. Then, the data fit to the hyperbolic binding isotherm weighted by the efficiency of crosslinking (β): [P•tRNA]/[tRNA] total = β X (1 / (1 + Kd /[P] total). The data were fit with the Kaleida Graph (Synergy Software, Reading, PA) curve-fitting program using the above equation.
Native and Temperature Gradient Gel Electrophoresis Assays
RNase P RNAs and cpRNAs were folded before native and temperature gradient gel electrophoresis experiments in 1X THE (66 mM HEPES, 33 mM Tris-HCl pH 7.4, 0.1 mM EDTA), 100 mM NH4OAc, 10 mM MgOAc and 0.05% NP40 for 10 min at 60°C followed by 20 min at 37 °C. For native gel Mg2+ titrations, samples were diluted 1:4 with loading buffer (1X THE, 13% sucrose, and .02% bromophenol blue) and ~ 200 nM RNA. The samples were loaded onto four 4.5% acrylamide 1X THE native gels containing 100 mM NH4OAc and various concentrations of MgOAc (0, 0.5, 1.0 and 5.0 mM). It was not necessary to dialyze the samples to the exact gel conditions because they equilibrated rapidly once in the gel. Electrophoresis running buffer contained 1X THE, 100 mM NH4OAc and the requisite MgOAc concentration. The running buffer was changed every hour. Each gel was run at constant voltage, 250 Volts, and detected by autoradiography or phosphorimager. TGGE gels samples were treated the same as above and the samples were loaded in 15 minute intervals in the single well. TGGE gels, buffers and running conditions were the same as above.
Computer Modeling
The crystal structure of B. stearothermophilus RNase P RNA, bacterial and eukaryal crosslinking data and the constraints of secondary structure were used to develop the S. pombe RNA tertiary structure model. Nonhomologous helices and single-stranded regions of S. pombe RNase P RNA were constructed and minimized individually using the Discover modules of Insight II program (Accelrys) and Crystallography and NMR System (CNS). The S. pombe RNA model was then further refined with energy minimization using CNS. The S. pombe RNA model was visualized using the modeling software by MacPyMOL DeLano, W.L.; The PyMOL Molecular Graphics System (2002) on the World Wide Web http://www.pymol.org
Supplementary Material
Acknowledgments
We thank Alexei Kazantsev for help in producing figures. We are grateful to Briana Dennehey and the entire Pace Lab for many helpful discussions. We thank Anne Gooding and the Cech Lab for the use of the TGGE equipment. We also thank Richard Maraia for helpful discussions about eukaryotic tRNAs. This work was supported by a grant from the NIH to NRP and NSF Alliance for Graduate Education and the Professoriate.
References
- Beebe JA, Kurz JC, Fierke CA. Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry. 1996;36:10493–10505. doi: 10.1021/bi960870m. [DOI] [PubMed] [Google Scholar]
- Brown JW. The Ribonuclease P Database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Nolan JM, Haas ES, Rubio MAT, Major F, Pace NR. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc Natl Acad Sci USA. 1996;93:3001–3006. doi: 10.1073/pnas.93.7.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin AB, Pace NR. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990;9:4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Pace NR. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Nolan JM, Harris ME, Pace NR. Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J. 1998;17:1515–1525. doi: 10.1093/emboj/17.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr SC, Zito K, Smith D, Pace NR. Contributions of phylogenetically variable structural elements to the function of the ribozyme ribonuclease P. Biochemistry. 1992;31:328–333. doi: 10.1021/bi00117a003. [DOI] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;22:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre A, Saintome C, Fourrey JL, Clivio P, Laugaa P. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid-protein interactions. J Photochem Photobiol B. 1998;42:109–124. doi: 10.1016/s1011-1344(97)00116-4. [DOI] [PubMed] [Google Scholar]
- Frank DN, Adamidi C, Ehringer MA, Pitulle C, Pace NR. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8:296–306. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Kazantsev AV, Chen JL, Pace NR. Analysis of the tertiary structure of the ribonuclease P ribozyme-substrate complex by site-specific photoaffinity crosslinking. RNA. 1997;3:561–576. [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Nolan JM, Malhotra A, Brown JW, Harvey SC, Pace NR. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J. 1994;13:3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Pace NR. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA. 1995;1:210–218. [PMC free article] [PubMed] [Google Scholar]
- Jarrous N. Human ribonuclease P: subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci U S A. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom LA. RNase P RNA-mediated catalysis. Biochem Soc Trans. 2002;30:1153–1158. doi: 10.1042/bst0301153. [DOI] [PubMed] [Google Scholar]
- Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem Biophys Res Commun. 2003;306:666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Marquez SM, Harris JK, Kelley ST, Brown JW, Dawson SC, Roberts EC, Pace NR. Structural implications of novel diversity in eucaryal RNase P RNA. RNA. 2005;11:739–751. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Shinitzky M, Smith RA. The activation of papain and ficin by phosphorothioate. Biochemistry. 1967;6:1421–1428. doi: 10.1021/bi00857a026. [DOI] [PubMed] [Google Scholar]
- Nolan JM, Pace NR. Structural analysis of the bacterial ribonuclease P RNA. In: Eckstein F, Lilley DMJ, editors. Nucleic Acids and Molecular Biology. Berlin: Springer-Verlag; 1996. pp. 109–128. [Google Scholar]
- Oh BK, Pace NR. Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994;22:4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BK, Frank DN, Pace NR. Participation of the 3′-CCA of tRNA in the binding of catalytic Mg2+ ions by ribonuclease P. Biochemistry. 1998;37:7277–7283. doi: 10.1021/bi973100z. [DOI] [PubMed] [Google Scholar]
- Pace NR, Brown JW. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J Bacteriol. 1995;177:1919–1928. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan-Ramos E, Lee Y, Engelke DR. Mutational analysis of Saccharomyces cerevisiae nuclear RNase P: randomization of universally conserved positions in the RNA subunit. RNA. 1996;2:441–451. [PMC free article] [PubMed] [Google Scholar]
- Pan T, Gutell RR, Uhlenbeck OC. Folding of circularly permuted transfer RNAs. Science. 1991;254:1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Pitulle C, Garcia-Paris M, Zamudio KR, Pace NR. Comparative structure analysis of vertebrate ribonuclease P RNA. Nucleic Acids Res. 1998;26:3333–3339. doi: 10.1093/nar/26.14.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich C, Olsen GJ, Pace B, Pace NR. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988;239:178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- Smith D, Burgin AB, Haas ES, Pace NR. Influence of metal ions on the ribonuclease P reaction. Distinguishing substrate binding from catalysis. J Biol Chem. 1992;267:2429–2436. [PubMed] [Google Scholar]
- Thomas BC, Kazantsev AV, Chen JL, Pace NR. Photoaffinity cross-linking and RNA structure analysis. Methods Enzymol. 2000;318:136–147. doi: 10.1016/s0076-6879(00)18049-8. [DOI] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- Tranguch AJ, Engelke DR. Comparative structural analysis of nuclear RNase P RNAs from yeast. J Biol Chem. 1993;268:14045–14055. [PubMed] [Google Scholar]
- Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- Zahler NH, Christian EL, Harris ME. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–745. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler WA, Morris J, Scott FH, Millikin C, Engelke DR. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


