Abstract
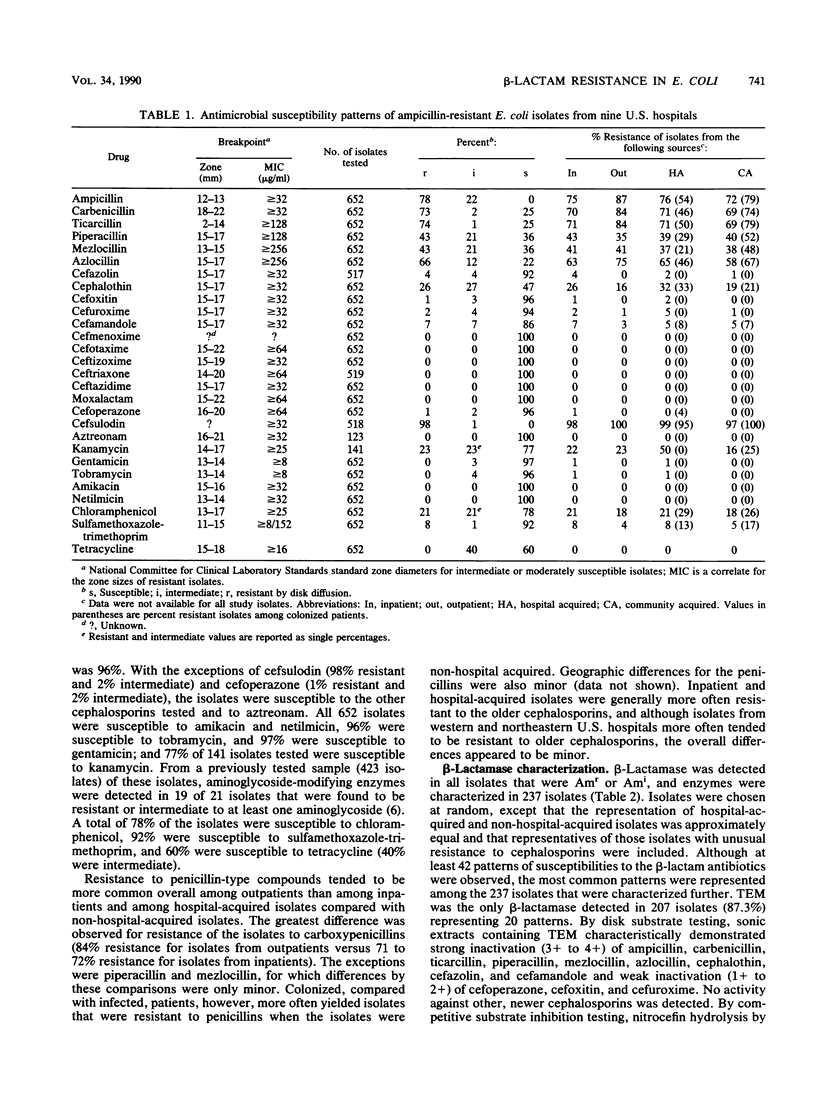
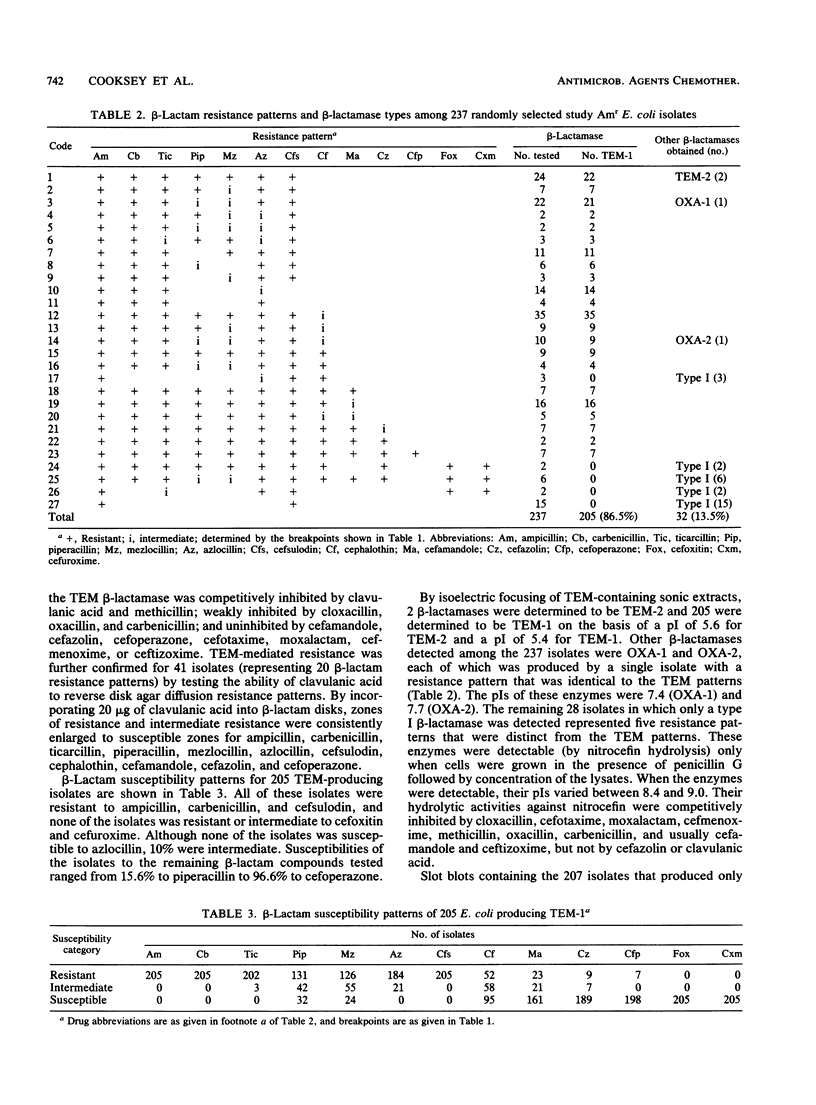
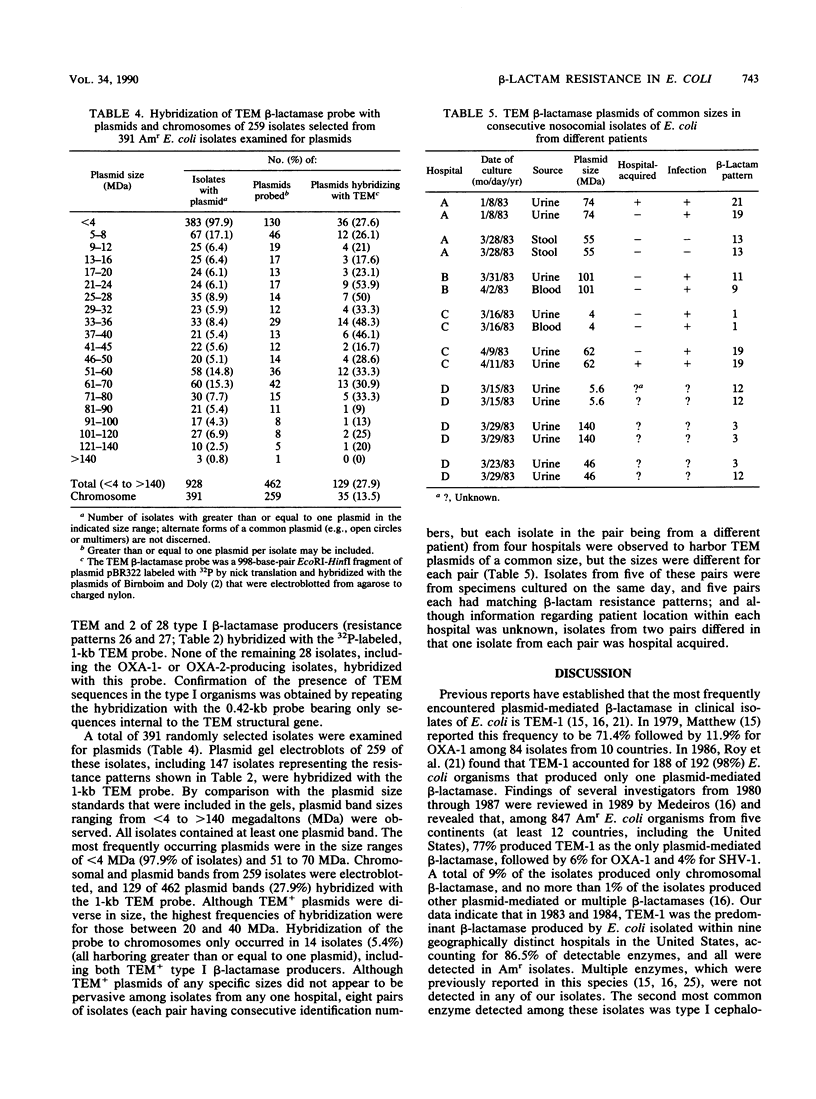
To study the national distribution of beta-lactam resistance patterns and mechanisms among Escherichia coli organisms isolated in U.S. hospitals, 652 ampicillin-resistant (Am(r)) or ampicillin-intermediate (Ami) isolates were submitted to the Centers for Disease Control from March 1983 through July 1984 by nine hospitals participating in the National Nosocomial Infections Study. Among the isolates (most of which caused urinary tract infections), 78% were Am(r) and 22% were Ami by the interpretative criteria established by the National Committee for Clinical Laboratory Standards. Resistance to carboxypenicillins ranged from 73 to 74%, and that to acylureidopenicillins ranged from 43 to 66%. A total of 26% of the isolates were resistant to cephalothin, and 4% were resistant to cefazolin. Resistance to cefoxitin was 1%, while resistances to cefuroxime and cefamandole were 2 and 7%, respectively. With the exception of cefsulodin (98% resistant) and cefoperazone (1% resistant), there was no resistance to newer cephalosporins or aztreonam. In general, only minor differences in the incidence of resistance to beta-lactam antibiotics were noted in hospital-acquired versus non-hospital-acquired isolates as well as among isolates from various regions of the United States. TEM beta-lactamases were produced by 87% of the 237 Am(r) isolates examined. By our methods, OXA and chromosomal (type I) beta-lactamases were detected in 2 and 28 isolates, respectively, and plasmid-mediated extended-spectrum cephalosporinases were detected in none of the isolates. Disk substrate and clavulanic acid inhibition assays revealed that TEM beta-lactamase conferred Am(r) and resistance to carboxypenicillins, acylureidopenicillins, cephalothin, cefamandole, cefsulodin, and cefoperazone. A total of 391 isolates were screened for plasmids, and 259 isolates were examined by DNA hybridization with a TEM probe. Among 462 plasmids probed, 129 plasmids, ranging from 4 to 140 megadaltons, harbored TEM sequences. Although beta-lactam resistance in clinical isolates of E. coli is predominantly mediated by TEM beta-lactamase, the diverse spectrum of resistance appears to be related to additional strain=dependent factors.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergström S., Normark S. Beta-lactam resistance in clinical isolates of Escherichia coli caused by elevated production of the ampC-mediated chromosomal beta-lactamase. Antimicrob Agents Chemother. 1979 Oct;16(4):427–433. doi: 10.1128/aac.16.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot M., Mercier J., Levesque R. C. Development of natural and synthetic DNA probes for OXA-2 and TEM-1 beta-lactamases. Antimicrob Agents Chemother. 1987 May;31(5):728–734. doi: 10.1128/aac.31.5.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey R. C., Clark N. C., Thornsberry C. A gene probe for TEM type beta-lactamases. Antimicrob Agents Chemother. 1985 Jul;28(1):154–156. doi: 10.1128/aac.28.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynes R. P., Cooksey R., Thornsberry C., Swenson J. M., Hughes J. M. Mechanism of aminoglycoside resistance among beta-lactam-resistant Escherichia coli in the United States. Diagn Microbiol Infect Dis. 1987 May;7(1):45–50. doi: 10.1016/0732-8893(87)90068-x. [DOI] [PubMed] [Google Scholar]
- Hiraoka M., Okamoto R., Inoue M., Mitsuhashi S. Effects of beta-lactamases and omp mutation on susceptibility to beta-lactam antibiotics in Escherichia coli. Antimicrob Agents Chemother. 1989 Mar;33(3):382–386. doi: 10.1128/aac.33.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovinen S., Huovinen P., Torniainen K., Jacoby G. A. Evaluation of plasmid-encoded beta-lactamase resistance in Escherichia coli blood culture isolates. Eur J Clin Microbiol Infect Dis. 1988 Oct;7(5):651–655. doi: 10.1007/BF01964244. [DOI] [PubMed] [Google Scholar]
- Huovinen S., Huovinén P., Jacoby G. A. Detection of plasmid-mediated beta-lactamases with DNA probes. Antimicrob Agents Chemother. 1988 Feb;32(2):175–179. doi: 10.1128/aac.32.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Sutton L. beta-Lactamases and beta-lactam resistance in Escherichia coli. Antimicrob Agents Chemother. 1985 Nov;28(5):703–705. doi: 10.1128/aac.28.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenot M., Deschaseaux M. L., Royez M., Mougin C., Cooksey R. C., Michel-Briand Y., Adessi G. L. Molecular hybridization versus isoelectric focusing to determine TEM-type beta-lactamases in gram-negative bacteria. Antimicrob Agents Chemother. 1987 Feb;31(2):300–305. doi: 10.1128/aac.31.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzis M. D., Billot-Klein D., Goldstein F. W., Williamson R., Tran Van Nhieu G., Carlet J., Acar J. F., Gutmann L. Dissemination of the novel plasmid-mediated beta-lactamase CTX-1, which confers resistance to broad-spectrum cephalosporins, and its inhibition by beta-lactamase inhibitors. Antimicrob Agents Chemother. 1988 Jan;32(1):9–14. doi: 10.1128/aac.32.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebe C., Nies B. A., Meyer J. F., Tolxdorff-Neutzling R. M., Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985 Aug;28(2):302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda G., Tomioka S., Hasegawa M. Detection of beta-lactamase production by gram-negative bacteria. J Antibiot (Tokyo) 1976 Jun;29(6):662–664. doi: 10.7164/antibiotics.29.662. [DOI] [PubMed] [Google Scholar]
- Matthew M. Plasmid-mediated beta-lactamases of Gram-negative bacteria: properties and distribution. J Antimicrob Chemother. 1979 Jul;5(4):349–358. doi: 10.1093/jac/5.4.349. [DOI] [PubMed] [Google Scholar]
- Normark S., Edlund T., Grundström T., Bergström S., Wolf-Watz H. Escherichia coli K-12 mutants hyperproducing chromosomal beta-lactamase by gene repetitions. J Bacteriol. 1977 Dec;132(3):912–922. doi: 10.1128/jb.132.3.912-922.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit A., Sirot D. L., Chanal C. M., Sirot J. L., Labia R., Gerbaud G., Cluzel R. A. Novel plasmid-mediated beta-lactamase in clinical isolates of Klebsiella pneumoniae more resistant to ceftazidime than to other broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1988 May;32(5):626–630. doi: 10.1128/aac.32.5.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy C., Segura C., Tirado M., Reig R., Hermida M., Teruel D., Foz A. Frequency of plasmid-determined beta-lactamases in 680 consecutively isolated strains of Enterobacteriaceae. Eur J Clin Microbiol. 1985 Apr;4(2):146–147. doi: 10.1007/BF02013586. [DOI] [PubMed] [Google Scholar]
- Sanders C. C., Iaconis J. P., Bodey G. P., Samonis G. Resistance to ticarcillin-potassium clavulanate among clinical isolates of the family Enterobacteriaceae: role of PSE-1 beta-lactamase and high levels of TEM-1 and SHV-1 and problems with false susceptibility in disk diffusion tests. Antimicrob Agents Chemother. 1988 Sep;32(9):1365–1369. doi: 10.1128/aac.32.9.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. C., Sanders W. E., Jr, Moland E. S. Characterization of beta-lactamases in situ on polyacrylamide gels. Antimicrob Agents Chemother. 1986 Dec;30(6):951–952. doi: 10.1128/aac.30.6.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot D., Chanal C., Labia R., Sirot J. Susceptibility of new beta-lactams to the expanded-spectrum beta-lactamase CTX-1. Infection. 1989 Jan-Feb;17(1):28–30. doi: 10.1007/BF01643496. [DOI] [PubMed] [Google Scholar]
- Sirot D., Sirot J., Saulnier P., Joly B., Chanal M., Cluzel M., Cluzel R. Resistance to beta-lactams in Enterobacteriaceae: distribution of phenotypes related to beta-lactamase production. J Int Med Res. 1986;14(4):193–199. doi: 10.1177/030006058601400405. [DOI] [PubMed] [Google Scholar]
- Sougakoff W., Goussard S., Gerbaud G., Courvalin P. Plasmid-mediated resistance to third-generation cephalosporins caused by point mutations in TEM-type penicillinase genes. Rev Infect Dis. 1988 Jul-Aug;10(4):879–884. doi: 10.1093/clinids/10.4.879. [DOI] [PubMed] [Google Scholar]


