Abstract
Resistance to the limited number of available antifungal drugs is a serious problem in the treatment of Candida albicans. We found that aneuploidy in general and a specific segmental aneuploidy, consisting of an isochromosome composed of the two left arms of chromosome 5, were associated with azole resistance. The isochromosome forms around a single centromere flanked by an inverted repeat and was found as an independent chromosome or fused at the telomere to a full-length homolog of chromosome 5. Increases and decreases in drug resistance were strongly associated with gain and loss of this isochromosome, which bears genes expressing the enzyme in the ergosterol pathway targeted by azole drugs, efflux pumps, and a transcription factor that positively regulates a subset of efflux pump genes.
Candida albicans is the most prevalent human fungal pathogen and is especially problematic in immune-compromised individuals. Azoles are antifungal drugs used extensively in the therapy of C. albicans infections because they cause few side effects. Resistance to azoles arises during long-term low-level prophylactic treatment regimes (1). The evolution of azole-resistance can occur via different pathways, e.g., increased activity of transcription factors that regulate drug pumps (2) or mutations in the ergosterol biosynthetic pathway (3), and may be facilitated by the heat shock protein Hsp90 (4). Although C. albicans does not have a complete sexual cycle, surveys of clinical strains suggest that it tolerates genome flexibility that generates a moderate level of selectable genetic diversity (5–11).
We adapted comparative genome hybridization (CGH) arrays for the analysis of gene copy number at all loci (10) for 70 azole-resistant and azole-sensitive strains from clinical and laboratory sources (table S1). We found 37 aneuploid chromosomes in 23 strains (Fig. 1A, black and white bars). Aneuploidy was seven times as prevalent in fluconazole-resistant [FluR, minimum inhibitory concentration (MIC) ≥ 4] (21 out of 42 strains) as in fluconazole-sensitive (FluS, MIC < 4) (2 out of 28 strains) isolates. Aneuploidy, primarily trisomy, was most prevalent on chr5 (15 events), which also exhibited a high level of segmental aneuploidy (8 events) (Fig. 1A, gray bars) (10).
Fig. 1.
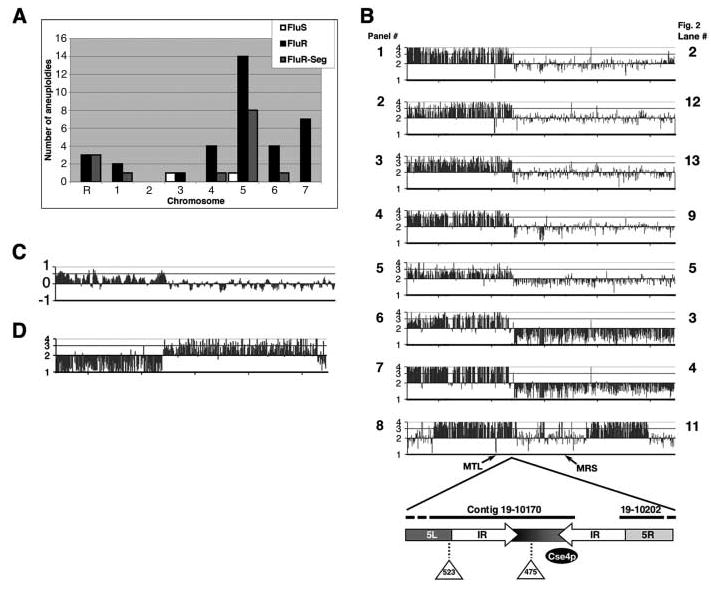
A common segmental aneuploidy appears in FluR strains. (A) Aneuploidy was common in FluR strains (table S1). Number of whole-chromosome aneuploidies in FluS (white) and FluR (black) and number of segmental aneuploidies, seen only in FluR strains (gray). (B) A common chr5 breakpoint in FluR strains. Panels 1 through 8 represent CGH plots of chr5 for YJB8735, YJB9185, YJB8640, YJB9311, YJB8738, YJB8736, YJB8737, and YJB9180. The y axis plots gene copy number calculated from log 2 values. Two shorter segmental aneuploidies are seen in panel 8. Diagram illustrates the fine structure of the breakpoint including the Cse4p site and the two insertions found on one homolog. IR, inverted repeat; MTL, mating type–like. (C) Increased gene copy number correlates with increased gene expression. Transcript profile data (mean log 2 values) for strain YJB9311 (26) were plotted by using a running average over five ORFs. (D) CGH data of chr5 from a FluS strain reveals an i(5R) with the same breakpoint as strains with i(5L).
All eight FluR strains with a chr5 segmental aneuploidy displayed distinctive features by CGH: increased gene copy number on the left arm of the chromosome; one or two copies of genes on the right arm of the chromosome; and, most strikingly, the breakpoint between the two arms was identical (Fig. 1B). This common breakpoint was located between open reading frames orf19.3161 and orf19.4219, which flank a gap between contigs 19.10170 and 19.10202 in the version 19 assembly of the genome sequence. We sequenced the region between these two contigs (fig. S1) from strains carrying either one of the two chr5 homologs, which allowed us to distinguish alleles found in this partially heterozygous region of the genome. The sequence gap was within one arm of an inverted repeat flanking the binding site for Cse4p, the histone H3 variant that is necessary for centromere function (12) (Fig. 1B, diagram).
The combination of CGH data and Southern blot analysis revealed an isochromosome (two identical chromosome arms flanking a centromere) in the eight strains with extra copies of chr5L. An isochromosome composed of two chr5L arms should include a unique fragment, spanning from one chr5L arm, through the inverted repeat and the centromere, to the other chr5L arm. This was verified by the presence of a unique ~10 thousand base pairs (kbp) Eco NI fragment (Fig. 2A) in all eight strains carrying the predicted isochromosome.
Fig. 2.
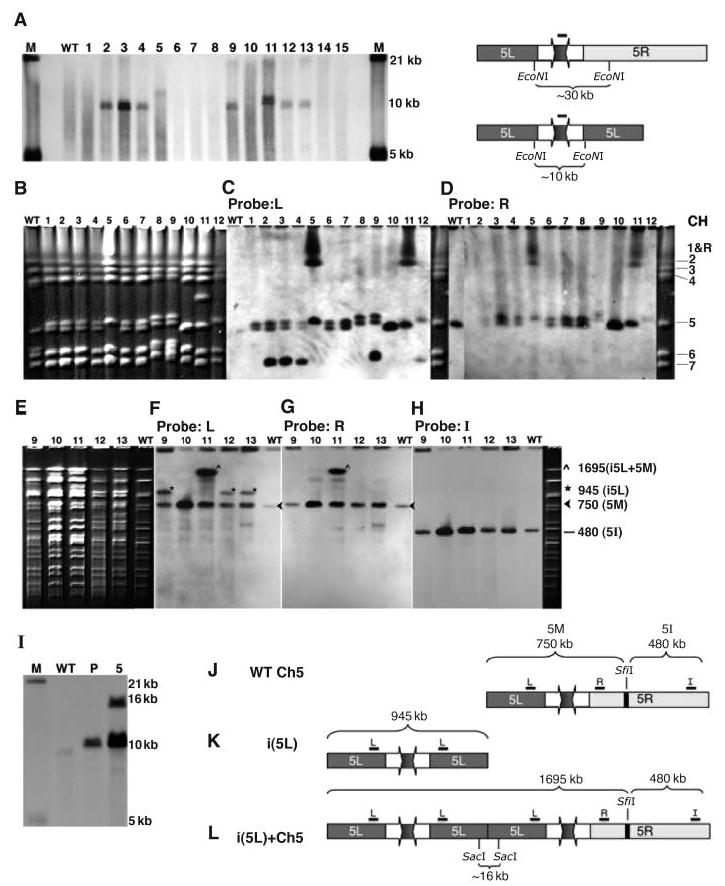
The segmental aneuploidies on chr5 reflect isochromosome structures. (A) All strains from Fig. 1B contain a unique restriction fragment that spans the Cse4p binding site (lanes 2 to 5, 9, and 11 to 13). Southern blot of Eco NI–digested genomic DNA probed with Cse4p binding site sequence (black bar) detects an ~10-kbp fragment diagnostic of i(5L) structure. Differences in the size of this fragment are due to polymorphic insertion sequences (Fig. 1B). WT: wild type (SC5314), lanes 1 to 15: YJB8734, YJB8735, YJB8736, YJB8737, YJB8738, YJB8739, YJB8740, YJB9309, YJB9311, YJB9175, YJB9180, YJB9185, YJB8640, YJB9613, and YJB8638. Lane designations are identical for all other parts of this figure. (B to H) The isochromosome is either independent or attached to a homolog of chr5. Whole-chromosome CHEF gels stained with ethidum bromide (B), blotted, and probed with chr5L (C) or chr5R (D) reveal an independent isochromosome (lanes 2 to 4, 9, and 12) or an attached isochromosome (lanes 5 and 11). Sfi I digestion of whole chromosomes separated by CHEF and stained with ethidium bromide (E); blotted; and probed with chr5L (F), chr5R (G), and chr5I (H) probes [diagramed in (J, K, and L)] show that the independent isochromosome (lanes 9, 12, and 13) does not contain an Sfi I site. Sfi I digestion of the attached isochromosome (lane 11) releases fragments expected if i(5L) were attached to the left arm of whole chr5. (I) The attached isochromosome includes a telomere-telomere junction. Southern blot analysis of Sac I–digested genomic DNA probed with a chr5L telomere-adjacent probe detected a 10-kbp telomere fragment in all wild-type (WT) and parental (P) strains and an additional larger fragment in YJB8738 (lane 5), as would be expected if two chr5L arms attached via telomere sequence (L). YJB9180 contains a complex attachment not including this Sac I fragment. (J to L) Diagrams, including relevant restriction sites and probes: (J) normal chr5, (K) independent i(5L), and (L) attached i(5L). The centromere region (inverted repeat and central unique sequence) is not to scale.
The predicted size of an independent chr5L isochromosome [i(5L)] is similar to that of a normal chr7. Consequently, ethidium bromide staining of whole-chromosome contour-clamped homogeneous electric field (CHEF) gels revealed no consistent changes in the karyotypes of most strains (Fig. 2B). Many strains had two separable chr5 homologs, likely due to differences in major repeat sequence (MRS) length in the two sister chromosomes (13), that were detected with probes from chr5L and chr5R (Fig. 2, C and D). Probes from chr5L, but not from chr5R, revealed an ~945-kbp band (twice the size of one chr5L arm) in six of the strains (Fig. 2C, lanes 2, 3, 4, 9, and 12). In these strains, the ~945-kbp band was not digested by the restriction enzyme Sfi I (Fig. 2, E to H), which cuts only within the MRS on the right arm of chr5 (14) (Fig. 2J).
Two other strains with chr5 segmental aneuploidy contained a larger band (~2.2 Mbp) that hybridized to probes from both chr5L and chr5R (Fig. 2, C and D, lanes 5 and 11) and only one chr5 homolog of normal size. Southern analysis of Sac I–digested DNA confirmed the presence of a novel, telomere-associated band in strain YJB8738 (Fig. 2I), suggesting that an isochromosome is attached to a full-length copy of chr5, by means of the left telomere (Fig. 2L). In this case, one of the centromeres must have been inactivated, either by a breakage and healing event (15) or by silencing of one of the centromeres (16).
Several genes on chr5L potentially have a role in FluR, including ERG11, which encodes lanosterol-14α-demethylase, the target of fluconazole (17), and TAC1, which encodes a transcription factor that up-regulates expression of adenosine triphosphate (ATP)–binding cassette (ABC) transporter genes on chr3 (18). In addition, there are genes encoding predicted efflux pumps: orf19.4144, which encodes an ABC transporter, and orf19.1942, which encodes a predicted multidrug resistance (MDR) transporter. Analysis of expression profiles confirmed that expression of most genes on chr5L were increased relative to expression of genes on chr5R in a strain carrying i(5L) (Fig. 1C), which may account for the increased FluR of these strains. Thus, as in Saccharomyces cerevisiae, chromosome copy number increases are associated with corresponding increases in expression of genes across the aneuploid region (19).
During a period of drug treatment, reversible resistance often develops such that when selection is relaxed, in vivo or in vitro, resistance levels can drop (20–22). The acquisition of unstable "hetero-resistance" has been documented in clinical isolates from a patient treated with azole antifungals (21). Strain YJB8638 (Fig. 3A) was the initial FluS isolate; strain YJB9185 was the isolate most resistant to fluconazole acquired from the same patient (Fig. 3B). This was an unstable strain that lost resistance when cultured in vitro in the absence of drug (22). The i(5L) aneuploidy was not present in the initial FluS isolate (Fig. 3A); appeared in the resistant strain (Fig. 3B); and subsequently was lost in YJB9517, a strain with significantly reduced FluR derived from YJB9185 in the absence of drug selection (Fig. 3C). All YJB9185-derived strains with dramatic reductions in FluR lost the segmental aneuploidy, whereas all strains that maintained high FluR also retained the segmental aneuploidy (table S3). YJB9185 and its derivatives are homozygous for a hyperactive allele of TAC1 (2), which may account for the residual FluR of YJB9517 relative to that of YJB8638 (Fig. 3). We cannot rule out an additional contribution from the short segmental monosomy on chr1 that appeared in YJB9185 and was retained in YJB9517 (Fig. 3).
Fig. 3.
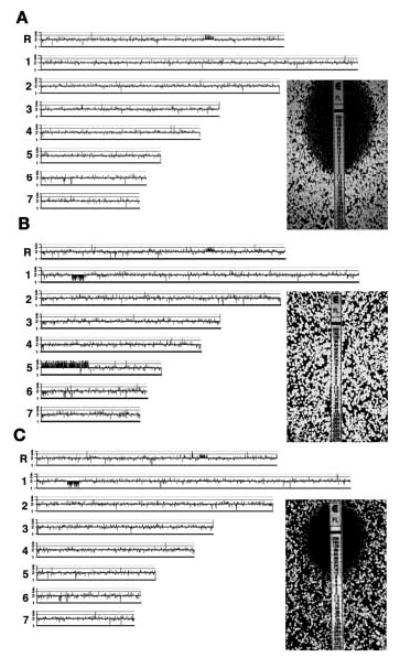
Loss of i(5L) is associated with a dramatic decrease in FluR. Whole-genome CGH plots reveal, compared with (A), the gain (B) and loss (C) of i(5L) (left panels) and fluconazole-resistance by E-test assay (right panels) in isolates: (A) YJB8638, MIC = 2.0 μg/ml; YJB9185, (B) MIC > 256 μg/ml; (C) YJB9517, MIC = 8 μg/ml.
Strain YJB8736, which evolved FluR in vitro, grew significantly better in the presence of fluconazole and significantly worse in the absence of fluconazole than the progenitor strain, which did not carry i(5L) (23). Under fluconazole selection, i(5L) was maintained through more than 165 generations in this strain (23, 24), which indicated that i(5L) can confer a growth advantage in the presence of azoles and can be maintained in the presence of drug and that its loss is accompanied by a decrease in FluR.
We have detected aneuploidies, primarily trisomies, for all C. albicans chromosomes (Fig. 1A) (10). This may reflect nondisjunction events like those that return tetraploids to a state where most, but not all, chromosomes are in the diploid state (8). Until now the i(5L) aneuploidy had not been detected because i(5L) migrates in CHEF gels with chr7, and the attached i(5L) migrates close to chr2.
Increased levels of gene expression from chr5L required for ergosterol biosynthesis and drug efflux pump activity are likely to contribute to drug resistance or tolerance. These genes do not occur on i(5R), and interestingly, aneuploidies involving this arm are not associated with drug resistance. It is noteworthy that the only sequence shared by the i(5L) and i(5R) isochromosomes, which thus delimits the chr5 centromere, is a 9.5-kbp region containing the unique sequence flanked by the inverted repeat (Fig. 1D). Consistent with the idea that replication forks pause at centromere sequences (25), we suggest that the centromere region of chr5 may be a fragile site where breaks occur and are healed more frequently.
C. albicans is an organism in which isochromosome formation has been found to occur in response to antifungal drug selection. Because i(5L) provides a selective growth advantage under drug conditions, C. albicans should provide a useful model system for studying the mechanisms that generate isochromosomes (and, in some cases, telomere-telomere attachments). Reagents that reduce chromosome breakage and/or recombination events resulting in isochromosome formation could be useful companions to current azole antifungal treatments.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/313/5785/367/DC1
Materials and Methods
Fig. S1
Tables S1 to S3
References and Notes
References
- 1.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. Antimicrob Agents Chemother. 2002;46:1704. doi: 10.1128/AAC.46.6.1704-1713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coste AT, et al. Genetics. 2006;172:2139. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson JB, et al. Genetics. 2003;163:1287. doi: 10.1093/genetics/163.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowen LE, Lindquist S. Science. 2005;309:2185. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 5.Forche A, Schönian G, Gräser Y, Vilgalys R, Mitchell TG. Fungal Genet Biol. 1999;28:107. doi: 10.1006/fgbi.1999.1164. [DOI] [PubMed] [Google Scholar]
- 6.Schonian G, et al. Mycoses. 1993;36:171. doi: 10.1111/j.1439-0507.1993.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 7.Tavanti A, et al. J Clin Microbiol. 2005;43:5601. doi: 10.1128/JCM.43.11.5601-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett RJ, Johnson AD. EMBO J. 2003;22:2505. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janbon G, Sherman F, Rustchenko E. Proc Natl Acad Sci USA. 1998;95:5150. doi: 10.1073/pnas.95.9.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selmecki A, Bergmann S, Berman J. Mol Microbiol. 2005;55:1553. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Magee BB, Dawson D, Magee PT, Kumamoto CA. Mol Microbiol. 2004;51:551. doi: 10.1046/j.1365-2958.2003.03852.x. [DOI] [PubMed] [Google Scholar]
- 12.Sanyal K, Baum M, Carbon J. Proc Natl Acad Sci USA. 2004;101:11374. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lephart PR, Chibana H, Magee PT. Eukaryot Cell. 2005;4:733. doi: 10.1128/EC.4.4.733-741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu WS, Magee BB, Magee PT. J Bacteriol. 1993;175:6637. doi: 10.1128/jb.175.20.6637-6651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser JA, et al. Eukaryot Cell. 2005;4:401. doi: 10.1128/EC.4.2.401-406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan BA, Blower MD, Karpen GH. Nat Rev Genet. 2001;2:584. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 17.White TC. Antimicrob Agents Chemother. 1997;41:1482. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. Eukaryot Cell. 2004;3:1639. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes TR, et al. Nat Genet. 2000;25:333. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 20.Calvet HM, Yeaman MR, Filler SG. Antimicrob Agents Chemother. 1997;41:535. doi: 10.1128/aac.41.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marr KA, White TC, van Burik JA, Bowden RA. Clin Infect Dis. 1997;25:908. doi: 10.1086/515553. [DOI] [PubMed] [Google Scholar]
- 22.Marr KA, et al. Antimicrob Agents Chemother. 1998;42:2584. doi: 10.1128/aac.42.10.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowen LE, Kohn LM, Anderson JB. J Bacteriol. 2001;183:2971. doi: 10.1128/JB.183.10.2971-2978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowen LE, et al. J Bacteriol. 2000;182:1515. doi: 10.1128/jb.182.6.1515-1522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenfeder SA, Newlon CS. Mol Cell Biol. 1992;12:4056. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. Antimicrob Agents Chemother. 2004;48:3064. doi: 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.We thank J. Anderson, P. T. Magee, P. D. Rogers, D. Sanglard, and T. White for providing strains and M. Gerami-Nejad, M. McClellan, M. Carlson, and J. Borton for technical assistance. We thank A. Nantel, M. van het Hoog, and P. T. Magee for genome map information; S. Bergmann for Chromosome_Map programming; and N. Springer, D. Kirkpatrick, J. Strathern, M. Kupiec, M. Basrai, and D. Sanglard for helpful discussions and review of the manuscript. This work was supported by NIH R01 AIO62427 to J.B., a Microbial and Plant Genomics Institute Integrative fellowship to A.S., and a microarray supplemental award DE10641-S to M. Edgerton. Sequence analysis of chr5 is deposited in GenBank, accession number DQ660981. The CGH array data are listed in GEO, the Gene Expression Omnibus, as series ‘‘CGH analysis of Candida albicans strains’’ with accession number GSE4995.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


