Abstract
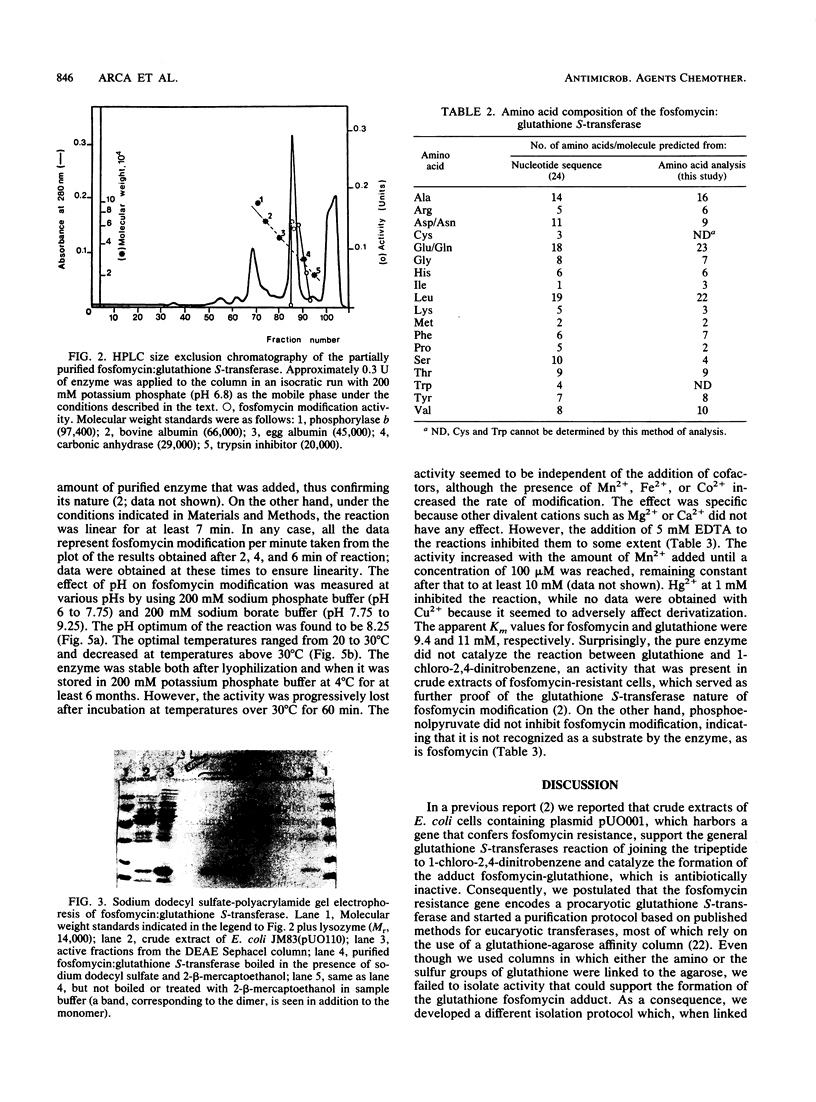
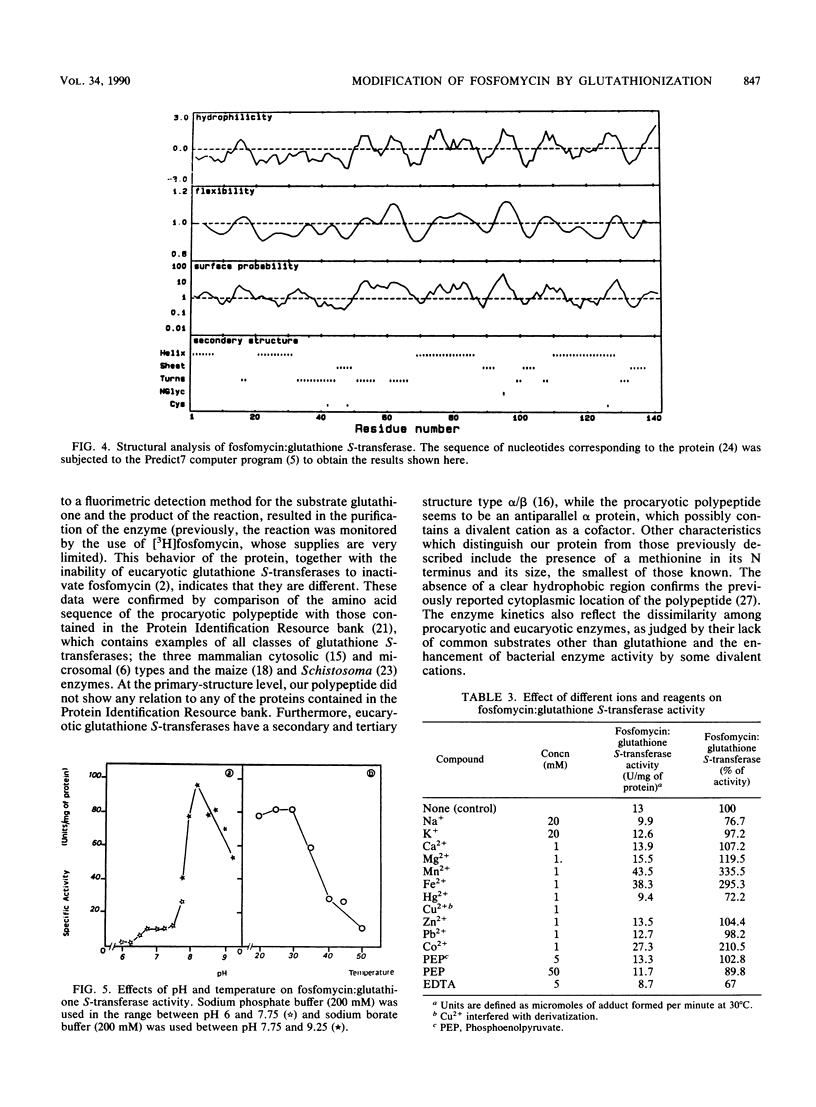
The enzyme that modifies fosfomycin by formation of an adduct with glutathione was purified 12-fold with a 56% activity yield by passage through DEAE Sephacel and high-performance liquid chromatography molecular exclusion columns. Its functional form was a homodimer of two 16,000-dalton polypeptides, which possibly showed an antiparallel alpha tertiary structure and which lacked marked hydrophobic regions. Visualization of the reaction was achieved by precolumn derivatization of glutathione and the adduct, separation by high-performance liquid chromatography, and fluorescence detection of both compounds. Temperature and pH optima were 20 to 30 degrees C and 8.25, respectively; Mn2+, Fe2+, and Co2+ enhanced the rate of modification; and Km values were 9.4 and 11 mM for fosfomycin and glutathione, respectively. Phosphoenolpyruvate did not interfere with fosfomycin modification. The enzyme was stable at 4 degrees C for at least 6 months but progressively lost its activity upon being heated for 60 min at temperatures over 30 degrees C.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arca P., Rico M., Braña A. F., Villar C. J., Hardisson C., Suárez J. E. Formation of an adduct between fosfomycin and glutathione: a new mechanism of antibiotic resistance in bacteria. Antimicrob Agents Chemother. 1988 Oct;32(10):1552–1556. doi: 10.1128/aac.32.10.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad D. W. Determination of D- and L-aspartate in amino acid mixtures by high-performance liquid chromatography after derivatization with a chiral adduct of o-phthaldialdehyde. Anal Biochem. 1984 Mar;137(2):405–409. doi: 10.1016/0003-2697(84)90106-4. [DOI] [PubMed] [Google Scholar]
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Cármenes R. S., Freije J. P., Molina M. M., Martín J. M. Predict7, a program for protein structure prediction. Biochem Biophys Res Commun. 1989 Mar 15;159(2):687–693. doi: 10.1016/0006-291X(89)90049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong J. L., Morgenstern R., Jörnvall H., DePierre J. W., Tu C. P. Gene expression of rat and human microsomal glutathione S-transferases. J Biol Chem. 1988 Jun 15;263(17):8430–8436. [PubMed] [Google Scholar]
- García-Lobo J. M., León J., Navas J., Ortiz J. M. Cloning and expression in minicells of the determinant of resistance to fosfomycin from the transposon Tn2921. Plasmid. 1984 May;11(3):243–247. doi: 10.1016/0147-619x(84)90030-1. [DOI] [PubMed] [Google Scholar]
- García-Lobo J. M., Ortiz J. M. Tn292l, a transposon encoding fosfomycin resistance. J Bacteriol. 1982 Jul;151(1):477–479. doi: 10.1128/jb.151.1.477-479.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Kadner R. J., Winkler H. H. Isolation and characterization of mutations affecting the transport of hexose phosphates in Escherichia coli. J Bacteriol. 1973 Feb;113(2):895–900. doi: 10.1128/jb.113.2.895-900.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan F. M., Kahan J. S., Cassidy P. J., Kropp H. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci. 1974 May 10;235(0):364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Alin P., Guthenberg C., Jensson H., Tahir M. K., Warholm M., Jörnvall H. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7202–7206. doi: 10.1073/pnas.82.21.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik B., Danielson U. H. Glutathione transferases--structure and catalytic activity. CRC Crit Rev Biochem. 1988;23(3):283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- Mannervik B. The isoenzymes of glutathione transferase. Adv Enzymol Relat Areas Mol Biol. 1985;57:357–417. doi: 10.1002/9780470123034.ch5. [DOI] [PubMed] [Google Scholar]
- Mendoza C., Garcia J. M., Llaneza J., Mendez F. J., Hardisson C., Ortiz J. M. Plasmid-determined resistance to fosfomycin in Serratia marcescens. Antimicrob Agents Chemother. 1980 Aug;18(2):215–219. doi: 10.1128/aac.18.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. E., Davies M. S., O'Connell K. M., Harding E. I., Wiegand R. C., Tiemeier D. C. Cloning and expression of a cDNA encoding a maize glutathione-S-transferase in E. coli. Nucleic Acids Res. 1986 Sep 25;14(18):7227–7235. doi: 10.1093/nar/14.18.7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas J., García-Lobo J. M., León J., Ortíz J. M. Structural and functional analyses of the fosfomycin resistance transposon Tn2921. J Bacteriol. 1985 Jun;162(3):1061–1067. doi: 10.1128/jb.162.3.1061-1067.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Sidman K. E., George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1988 Mar 11;16(5):1869–1871. doi: 10.1093/nar/16.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons P. C., Vander Jagt D. L. Purification of glutathione S-transferases by glutathione-affinity chromatography. Methods Enzymol. 1981;77:235–237. doi: 10.1016/s0076-6879(81)77031-9. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Davern K. M., Board P. G., Tiu W. U., Garcia E. G., Mitchell G. F. Mr 26,000 antigen of Schistosoma japonicum recognized by resistant WEHI 129/J mice is a parasite glutathione S-transferase. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8703–8707. doi: 10.1073/pnas.83.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruoka T., Yamada Y. Charactertization of spontaneous fosfomycin (phosphonomycin)-resistant cells of Escherichia coli B in vitro. J Antibiot (Tokyo) 1975 Nov;28(11):906–911. doi: 10.7164/antibiotics.28.906. [DOI] [PubMed] [Google Scholar]
- Venkateswaran P. S., Wu H. C. Isolation and characterization of a phosphonomycin-resistant mutant of Escherichia coli K-12. J Bacteriol. 1972 Jun;110(3):935–944. doi: 10.1128/jb.110.3.935-944.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar C. J., Hardisson C., Suárez J. E. Cloning and molecular epidemiology of plasmid-determined fosfomycin resistance. Antimicrob Agents Chemother. 1986 Feb;29(2):309–314. doi: 10.1128/aac.29.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



