Abstract
A central problem in science is deciding among competing explanations of data containing random errors. We argue that assessing the “complexity” of explanations is essential to a theoretically well-founded model selection procedure. We formulate model complexity in terms of the geometry of the space of probability distributions. Geometric complexity provides a clear intuitive understanding of several extant notions of model complexity. This approach allows us to reconceptualize the model selection problem as one of counting explanations that lie close to the “truth.” We demonstrate the usefulness of the approach by applying it to the recovery of models in psychophysics.
How does one decide among competing explanations of data, given limited observations? This problem of model selection is at the core of progress in science. It is particularly vexing in the statistical sciences, where sources of error are diverse and hard to control. For example, in psychology experiments, the participants are themselves a serious source of uncontrolled random variation. Choosing between candidate models that purport to describe underlying regularities about human behavior given noisy data is correspondingly problematic. Over the decades, scientists have used various statistical tools to select among alternative models of data but have lacked a clear theoretical framework for understanding model selection. The purpose of this article is to alert scientists to the importance of accounting for complexity when choosing among models and to provide a geometric formulation of complexity. Not only does a geometric approach recast model selection in a more intuitive and meaningful light, but it also provides insight into the relations among conventional statistical techniques and the inherent tradeoffs between model performance and complexity.
Statistical Model Selection: Issues and Problems.
From a statistical standpoint, the data are a sample generated from a true but unknown probability distribution, which is the regularity underlying the data. A statistical model is a parametric family of probability distributions defined on random variables Y, representing experimental data. Repeated measurement of Y yields some empirical distribution of observed values yi, which is presumed to tend to some truth t(yi) that the experiment seeks to uncover. The truth is statistically modeled in terms of a parameter vector θ, and a family of distributions f(yi|θ), which may not actually include the truth t. The scientist seeks to construct the best model of t within the family f.
This paradigm can model many different experimental situations. For example, some random variables may be controlled by the experimentalist, who sets the value of these quantities (X) and measures the distribution on the remaining Y. In this case, we speak of a dependent variable Y and an independent variable X, and the experiment probes the set of conditional distributions Pr(Y|X). If the scientist selects X according to the distribution Pr(X), the joint distribution Pr(Y, X) = Pr(Y|X) Pr(X) is the truth the experiment seeks, which can be modeled as above. In some circumstances, Y has a deterministic component as a function of X and a fixed random error piece. In that case, it is convenient to write the observed value of the dependent variable as yi = h(θ, xi) + ei (i = 1, … , N). Here h(θ, xi) is the mean of yi given a particular value (xi) of an independent variable X, θ is the parameter vector of the model, ei is an “error” that is distributed with zero mean, and N is the sample size.‡ We will study a general formalism for statistical inference of truths t(yi) with parametric model families f(yi, θ) and will illustrate our results with examples containing dependent and independent variables.
The goal of statistical model selection is straightforward: given a set of observations (data) corrupted by noise, select from a set of competing explanations the model that best captures the data's regularities. Achieving this goal is not straightforward because of the difficulty in reconciling two desirable yet conflicting properties of a good model: generalizability and goodness of fit.
Goodness of fit refers to how well a model fits a particular pattern of observed data. It is measured by comparing predicted outcomes of a model, with optimized parameter values, against the observed data. For example, the mean squared error (MSE) discrepancy measure is often used. Generalizability refers to how well a model, inferred on the basis of a set of observed data, predicts the statistics of new, as yet unseen, samples. In other words, suppose the model is fitted to the initial set of data. If the model—instantiated with the parameter values fitted to the initial set of data—also gives a good fit to future data samples drawn from the same underlying distribution or regularity, we say that it generalizes well. It can be difficult to define what this means formally, and many different but related measures of generalizability appear in the literature. Sometimes one directly measures proximity of an inferred model to a known truth by using a metric on the space of distributions (see, e.g., ref. 1 and references therein). When a true distribution t exists, generalizability may be defined in terms of a discrepancy measure Err(Data|Model),§ as the expected error in predicting future data given the inferred model: EtErr(Data|Model). Another notion of generalizability, used in the information theoretic literature, relates the length of codes obtained for data by using the sample statistics predicted by an inferred model [see, e.g., the seminal paper of Rissanen (2)], to be discussed below. All these methods of assessing generalizability are different, yet related: all measure prediction of future data statistics but yield somewhat different quantitative convergence rates to the best model. In the practical examples studied here, we will simply estimate generalizability by comparing inferred models, with parameter values fitted to initial data, to additional samples drawn randomly from a known truth. The best generalizing model is the one with the best fits to the additional data.
A central goal of statistical inference is to select the model that generalizes best and thus gives the best description of the underlying regularity. However, the very features of a model that improve goodness of fit to observed data can decrease generalizability (see, e.g., ref. 3). The following example illustrates this relationship.
Goodness of fit and generalizability are influenced by the number of parameters and the model's functional form (how the parameters are combined in the model equation). Together they contribute to a model's complexity, a concept that connotes the flexibility inherent in a model that enables it to fit diverse patterns of data. In Table 1, we compare the ability of three models to fit two data samples generated by one of the models (M1). Each model's parameters are chosen to give the best fit to the first sample. With these parameters fixed, generalizability is assessed by fitting to the second sample.
Table 1.
Goodness of fit and generalizability of models differing in complexity
| Model | M1 (true model) | M2 | M3 |
|---|---|---|---|
| Goodness of fit | 4.28 (0%) | 3.84 (25%) | 3.67 (75%) |
| Generalizability | 5.37 (59%) | 5.62 (23%) | 5.78 (18%) |
Mean squared error (MSE) of the fit of each model to the data and the percentage of samples in which the particular model fitted the data best (in parentheses). The three models were as follows: M1: y = θ0 + θ1x + error, M2: y = θ0 + θ1xθ2 + θ3exp(x) + error, and M3: y = θ0 + θ1x + θ2x2 + θ3x3 + error. The error was normally distributed with a mean of zero and a standard deviation of 5. One thousand pairs of samples were generated from the model M1 using (θ0 = 1, θ1 = 2) on the same 10 points for x, which ranged from 0.1 to 4.6 in increments of 0.5.
The first row of Table 1 shows each model's mean fit to data drawn from M1, M2 and M3, with two more parameters than M1, always fit the data better than M1 itself. The improved fit relative to M1 (the true model) occurs because the two extra parameters in M2 and M3 allow them to absorb random error in the data. The latter models have therefore overfitted the data beyond what is necessary to capture the underlying truth. Furthermore, M3 fits better than M2. This improvement must be due to functional form, because these two models differ only in how parameters and data are combined in the model equation.
Results in the second row of Table 1 demonstrate that poor generalizability is the cost of overfitting a specific data sample. Not only are MSEs now greater for M2 and M3 than for M1, but both models provide the best fit to the second sample much less often than M1.
This example illustrates that the best-fitting model does not necessarily generalize the best. The trademark of a good model selection procedure is its ability to satisfy these two opposing goals. We desire a model that is complex enough to describe the data sample accurately but without overfitting and thus losing generalizability. To this end, a quantitative measure of complexity must account for both the number of parameters and the functional form of a model. In other words, we would like an analytic realization of Occam's Razor.
Previous Approaches to Measuring Model Complexity.
The overarching goal of many model selection approaches has been the estimation of a model's generalizability (for a review, see ref. 4). Some representative methods used for inference of parametric models are the Akaike Information Criterion [AIC, (5)], the Bayesian Information Criterion [BIC, (6)] and Rissanen's Stochastic Complexity [SC, (2, 7)]:
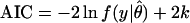 |
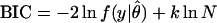 |
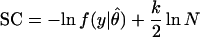 |
Here y = (y1, … , yN) is a data sample of size N, ln f(y|θ̂) is the maximized logarithm likelihood of the data y given the model, θ̂ is the maximum likelihood parameter estimate, and k is the number of parameters. The first term, which is similar across criteria, represents lack of fit to the data sample. The remaining term represents model complexity. The model minimizing one of these criteria is expected to generalize best and should be chosen.
For very small sample sizes, these model selection criteria may require careful interpretation and application. Originally, all of them were derived as approximations, for large sample sizes, of more general quantities that are hard to compute but more readily applicable to inference from small samples. For example, in a later section, we will show that SC arises as an asymptotic approximation to the likelihood that a parametric family contains the truth, given an observed collection of data. Therefore, for very small sample sizes, the complete likelihood should guide model selection.
Barron and Cover have shown that a large class of such Minimum Complexity Density Estimation (MCD) methods are consistent (1) (i.e., if the true model lies in the model space, it is recovered in the limit of large sample sizes). This provides a strong indication that these methods, at least asymptotically, generalize well. In the present context, define a model selection criterion MCD = −ln f(y|θ̂) + L(f), where L(f) is any reasonable measure of the complexity of the model family f, including Kolmogorov complexity (the length of the shortest computer program required to describe f on a universal Turing machine) (1, 8). The only restriction on the set {L(f)} for the models under consideration is that its elements must satisfy certain inequalities (1).¶ Our purpose in this article is to discuss the choice of L(f), the measure of model complexity. AIC, BIC, and SC are the most commonly used selection methods, but they include only the number of parameters (k) and the sample size (N). This seems inadequate because, as illustrated in Table 1, the functional form of a model is also pertinent to its generalizability. SC (9) improves on this shortcoming. However, it fails to meet the crucial requirement of being invariant under reparametrization of the model, a condition that any meaningfully interpretable measure of complexity must satisfy. From a statistical standpoint, the parameters simply index the collection of distributions a model describes; thus, the choice of parametrization should be irrelevant (10).
One way of determining a good complexity measure L(f) is the Minimum Description Length principle (MDL), which states that the best model for describing a set of data is the one that permits the greatest compression of the data description. This idea originated in algorithmic coding theory (8, 11, 12) with the notion that the existence of underlying regularities governing a collection of data necessarily implies redundancy in the information gained from successive observations (see ref. 13 for a review). That is, the more we compress data by extracting redundancy from it, the more we learn about underlying regularities (see, for example, refs. 14 and 15).
With this in mind, Rissanen has proposed the following reparametrization-invariant modification of his SC criterion (16):
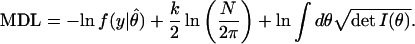 |
Here Iij(θ) is the Fisher Information matrix, defined as the expectation value Iij(θ) = −Eθ[∂2ln f(y|θ)/∂θi∂θj] evaluated in the distribution indexed by θ with a sample of size 1. The last two terms in MDL should be seen as the intrinsic complexity of the model family f. Rissanen proves that as N grows large, MDL is the length in bits of the shortest possible code describing data generated by a model lying within the family f. This suggests that the model that minimizes MDL uncovers the greatest amount of underlying regularity in the data. As such, the models it selects should perform well under any reasonable measure of generalizability, including the expected-future-error criterion discussed earlier. The same MDL criterion arises as an asymptotic approximation to the Bayesian posterior probability of a model given the data, for a special form of the parameter prior density (17). Hence, MDL minimization corresponds to maximization of the posterior probability within a Bayesian framework.
MDL is within the class of inference schemes to which Barron and Cover's results on Minimum Complexity Density Estimation apply (some work is necessary to cast the formalism of this paper into the language of Propositions 1–4 in ref. 1). So MDL will eventually recover the truth if it lies within a model family under consideration. Proposition 4 of ref. 1 bounds the rate at which the inferred model will converge to the truth. This translates into a bound on the generalization error of the inferred model. When the truth lies outside the considered models, analytical proofs regarding generalization error are hard to obtain under general conditions. However, for reasons presented below, one expects that MDL nevertheless selects models that come “close” to the truth and thereby generalize well.
MDL represents important progress in defining complexity and tackling the model selection problem. In practice, however, complexity has been conceptualized primarily as some combination of the number of parameters in a model and its functional form. This mode of thought can be misleading and can result in idiosyncratic heuristic solutions to the model selection problem. In this paper, we argue that various well-founded approaches to complexity such as MDL and Bayesian model selection can be understood elegantly and intuitively from a geometric perspective.
A Geometric Approach to Complexity
The Geometry of Parameter Manifolds.
From a geometric perspective, a parametric model family of probability distributions forms a Riemannian manifold embedded in the space of all distributions. Every distribution is a point in this space, and the collection of points created by varying the parameters of the model gives rise to a hypersurface in which “similar” distributions are mapped to “nearby” points.‖ The small distance ds between points separated by infinitesimal parameter differences dθi is given by ds2 = ∑i,j=1kgij(θ)dθi dθj, where gij(θ) is a Riemannian metric tensor. It has been argued from a variety of perspectives (10, 17–24), that the Fisher Information defined earlier is the natural metric on a manifold of distributions, that is, gij(θ) = Iij(θ).
Earlier we described complexity as the characteristic of a model
that enables it to fit a wide range of data patterns. In the present
context, complexity should be that characteristic of a model that
enables it to describe a wide range of probability distributions.
Models that describe “more” distributions should be more complex.
In effect, the complexity of a model family f should relate
to the volume the associated manifold occupies in the space of
distributions. The infinitesimal volume of the little cube formed by
the dθi is given by dV ≡
Πi=1k dθi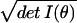 ≡
dθ
≡
dθ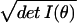 and is known as the
Riemannian volume element in differential geometry. The
volume of the model family f is then V(f) = ∫
dθ
and is known as the
Riemannian volume element in differential geometry. The
volume of the model family f is then V(f) = ∫
dθ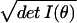 , where we integrate over the
entire parameter manifold. [We will always cut off the ranges of
parameters to ensure that V(f) is finite. These ranges
should be considered part of the functional form of the model.] By
construction, this volume is independent of the parametrization.
, where we integrate over the
entire parameter manifold. [We will always cut off the ranges of
parameters to ensure that V(f) is finite. These ranges
should be considered part of the functional form of the model.] By
construction, this volume is independent of the parametrization.
To recap, if we accept the Fisher Information as the natural metric on a parameter manifold, the volume V(f) can be seen as a quantity related to model complexity. We will now explain the exact nature of this relation by clarifying, (i) what we mean by the volume of a family of distributions, and (ii) why the above dV is the “natural” volume integration measure on a model manifold.
Determining the volume measure dV requires counting the distributions within the infinitesimal parameter volume Πidθi. In a statistical context, two distributions are indistinguishable if it is difficult to guess which of the two generated a typical large set of data drawn from either. Because of the way a model family is embedded in the space of distributions, two different parameter values can index very similar distributions. Our goal is to devise a measure of volume that counts only distinguishable distributions.
To achieve this goal, imagine playing an inference game: draw
data from one distribution, say θp, in the
model and ask how well we can guess whether the data came from
θp rather than from a nearby
θq. Our ability to distinguish between these
distributions increases with the amount of available data. However, it
is shown in the Appendix that for any fixed amount of data
there is a little ellipsoid around θp where
the probability of error in the guessing game is large. In other words,
within this ellipsoid, distributions are not very different in the
statistical sense. To count distinguishable distributions, we should
cover the manifold with such ellipsoids, counting one distribution for
each ellipsoid. This discretizes the parameter manifold to become a
lattice. We then want to take the limit of infinite sample size so that
the ellipsoids of indistinguishability shrink, and the associated
lattice becomes finer, forming a continuum in the limit. Taking this
limit recovers a continuum integration measure that counts only
distinguishable distributions. Strictly speaking, any finite coordinate
volume will contain an infinite number of distinguishable
distributions. However, as shown in the Appendix, the
ratio of distinguishable distributions contained in any two
regions V1 and V2 is
∫V1 dθ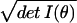 /∫V2 dθ
/∫V2 dθ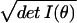 . It will transpire that
complexity is related to such ratios, which are assessed using
dV = dθ
. It will transpire that
complexity is related to such ratios, which are assessed using
dV = dθ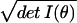 as the volume
integration measure on the parameter manifold (see refs. 10 and 17 for
details).
as the volume
integration measure on the parameter manifold (see refs. 10 and 17 for
details).
In summary, dV gauges the number of different
distributions indexed within an infinitesimal volume. (More precisely,
dV/∫ dθ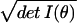 measures
the fraction of the distributions in the parametric family that are
contained in dV.) In effect, the
indistinguishable distributions have been “divided
out” of the integration measure. This makes good sense in the
context of measuring complexity: if complexity is related to the volume
of a model in the space of distributions, the measure of volume should
include only different, or distinguishable, distributions, and not the
artificial coordinate volume.
measures
the fraction of the distributions in the parametric family that are
contained in dV.) In effect, the
indistinguishable distributions have been “divided
out” of the integration measure. This makes good sense in the
context of measuring complexity: if complexity is related to the volume
of a model in the space of distributions, the measure of volume should
include only different, or distinguishable, distributions, and not the
artificial coordinate volume.
We have built a notion of volume of a parameter manifold from the
continuum limit of a counting of probability distributions. It is worth
remembering that, whereas “counts” can be compared across model
families of different dimension, volumes cannot. As an example,
consider a model A with parameters {θ1,
θ2}, having ranges {0 ≤
θ1 ≤ r}, {0 ≤ θ2 ≤
r} and a Fisher Information equal to the identity matrix.
Consider a family B containing distributions in family A with
parameters {θ1, 0}. Then the volume of model A is
VA = ∫ dθ1
dθ2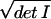 = r2,
whereas the volume of model B is VB = r.
For small r, VB is numerically greater than
VA, although every distribution in B is
manifestly contained in A also. On the other hand, if we discretize the
parameters with a grain size of ɛ < r, the
numbers of lattice cells within A and B are
NA = (r/ɛ)2 and
NB = r/ɛ, respectively. These may be
meaningfully compared and, for any r,
NA/NB = r/ɛ > 1, indicating
that A always contains more cells than B. Here
r2/ɛ2 and r/ɛ are
ratios of the total volumes of A and B and the volumes of their
discretized lattice cells. Similarly, any ratio of volumes
can be meaningfully compared across models of different dimension. As
we will see below, complexity of a model, from the MDL and Bayesian
perspectives, is related to the ratio of the total volume of the model
to the volume occupied close to the truth.
= r2,
whereas the volume of model B is VB = r.
For small r, VB is numerically greater than
VA, although every distribution in B is
manifestly contained in A also. On the other hand, if we discretize the
parameters with a grain size of ɛ < r, the
numbers of lattice cells within A and B are
NA = (r/ɛ)2 and
NB = r/ɛ, respectively. These may be
meaningfully compared and, for any r,
NA/NB = r/ɛ > 1, indicating
that A always contains more cells than B. Here
r2/ɛ2 and r/ɛ are
ratios of the total volumes of A and B and the volumes of their
discretized lattice cells. Similarly, any ratio of volumes
can be meaningfully compared across models of different dimension. As
we will see below, complexity of a model, from the MDL and Bayesian
perspectives, is related to the ratio of the total volume of the model
to the volume occupied close to the truth.
Inference and Geometry.
A model containing many distributions that come “close” to the truth that generated the data should give a good description of it. Proximity to the truth is assessed in probability theory by asking: What is the likelihood that the truth lies in the model family f given the observed data y? The Bayesian method for evaluating this likelihood uses the inversion rule:
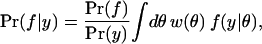 |
1 |
where Pr(f), Pr(y), and w(θ) are the a priori probabilities of the model family f, the data y, and the parameter value θ, whereas f(y|θ) is the probability that the observed data were generated by the model indexed by θ. In the absence of prior knowledge, Bayesian methods say that all model families and distributions should be considered equally likely. This involves neglecting Pr(f) and requires the prior distribution w(θ) to give equal weight to each distinguishable distribution indexed by the model parameters. Fortunately, we have solved the problem of counting distinguishable distributions and can set
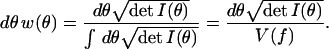 |
2 |
Because Pr(y) is a fixed number, Bayesian analysis says that the most likely model family given the data y is the one that maximizes
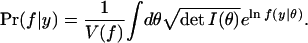 |
3 |
(Again, we have dropped the prior probability of f and y, which are the same across models and therefore appear as irrelevant constants.) We will show that choosing models with high Pr(f|y) is essentially equivalent to an MDL model selection criterion and will then interpret both methods from a geometric perspective.
As the number of data points N = |y| grows, the integrand in Pr(f|y) becomes peaks sharply around the parameter θ̂, which maximizes the likelihood f(y|θ). So, in the vicinity of θ̂, the integrand will be well approximated by a multivariate Gaussian with a covariance matrix N Jij(θ̂), where Jij(θ̂) = −(1/N) [∂2ln f(y|θ)/∂θi∂θj]θ=θ̂.** (We have separated the factors of N in this way because (1/N) ln f(y|θ) approaches a finite limit as N → ∞.) When N is large, the Gaussian is very narrow, and the integral can be performed to give Pr(f|y) ≈ f(y|θ̂)(Vc(f)/V(f)). We have defined (10, 17):
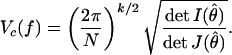 |
Vc(f) is essentially the volume of a small ellipsoid around θ̂ within which the probability of the data f(y|θ) is appreciable. Specifically, Vc(f) differs only by a numerical factor from the volume of a region where f(y|θ) ≥ λ f(y|θ̂) for any λ close to 1. As such, it measures the volume of distinguishable distributions in f that come close to the truth, as measured by predicting the data y with good probability. It is shown in refs. 10, 17, and 25 that there is a systematic expansion
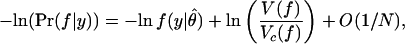 |
where the omitted terms vanish as N → ∞. The ratio Vc(f)/V(f) penalizes models that occupy a small volume close to the truth relative to the total volume of the model. The second term expands to
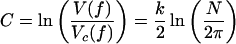 |
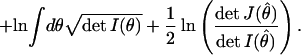 |
In Bayesian model selection, C functions as a penalty for complexity. We will analyze the meaning of this complexity in geometric terms and relate it to the classic stochastic complexity and MDL model selection criteria of Rissanen.
The first and second terms in C are independent of the true distribution as well as the data and therefore represent an intrinsic property of the model family. We will call them the geometric complexity of the model. The third term, which depends on the data, measures the complexity of the description of data drawn from a given true distribution, and so we call it the relative complexity. By construction, both geometric and relative complexity are invariant under reparametrization of the model. Moreover, if the true distribution lies within the considered model family, J(θ̂) approaches I(θ̂) as N grows large, and consequently, ln(V(f)/Vc(f)) becomes equal to the complexity penalty in the MDL selection criterion. This shows that as the sample size grows, the log of the Bayesian posterior probability of a model family [−ln Pr(f|y)] coincides with MDL when the truth lies in the model family. Therefore, selecting the most probable model is essentially equivalent to choosing the model that gives the MDL of the data, and the Bayesian complexity C coincides with Rissanen's modified SC. Both are equal to the quantity we are calling geometric complexity. In the calculation of the geometric complexity, the determinant of the Fisher Information matrix can be singular, meaning that its value becomes infinite at certain values of the parameter. When this occurs, parameter ranges may have to be restricted to ensure that the integral of the determinant remains finite.
Relative complexity is important when the truth lies outside the model family and measures the robustness of the model (its sensitivity to the precise choice of parameters) (10, 17). Note that the first term in C increases logarithmically with sample size (N), whereas the second and third terms are independent of N. This means that as N grows large, the effects of complexity because of functional form [I(θ)] will gradually diminish compared with those due to the number of parameters (k), thereby reducing the entire complexity measure to that of BIC.
Finally, we arrive at an intuitive geometric interpretation of the meaning of complexity in the MDL and Bayesian approaches to model selection: “complexity” measures the ratio of the volume occupied by distinguishable distributions in a model that come close to the truth relative to the volume of the model as a whole. Interpreting the relative volume ratio V/Vc in terms of the actual number of distributions contained within V and Vc requires care. Strictly speaking, the volumes V and Vc each contain an infinite number of distributions, but their finite ratio is interpreted as the fraction of distributions lying close to the truth. Therefore, a complex model is one with a small fraction of its distinguishable probability distributions lying near the truth. It is natural that MDL depends only on a volume ratio such as V/Vc because these can be compared across models with different numbers of parameters, unlike the volumes themselves.
Interestingly, the leading MDL penalty for model dimension,
(k/2) ln(N), has its geometrical origin in the
scaling of volumes with radius in different dimensions. Examination of
the Bayesian posterior shows that the integrand is appreciable in a
region of parameters around the maximum likelihood where |θ −
θ̂| is O(1/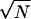 ). In k
dimensions, the volume of such a region diminishes as
(1/N)k/2. The resulting scaling of
Vc with N penalizes high
dimensionality. Higher dimensional models will generally contain
“more” probability distributions and may therefore better fit
statistical fluctuations in data. However, we are seeing here that
better fits are achieved at the cost of decreased robustness in the
choice of parameters; namely, the precision required in the parameters
to achieve a given degree of fit increases rapidly with dimensionality.
In this way, the simple scaling of volumes with radius in k
dimensions translates into a complexity penalty in the MDL and Bayesian
model selection methods. [The constant term (k/2)ln(2π)
has its origins in the details of the analysis and is less easy to
explain intuitively.]
). In k
dimensions, the volume of such a region diminishes as
(1/N)k/2. The resulting scaling of
Vc with N penalizes high
dimensionality. Higher dimensional models will generally contain
“more” probability distributions and may therefore better fit
statistical fluctuations in data. However, we are seeing here that
better fits are achieved at the cost of decreased robustness in the
choice of parameters; namely, the precision required in the parameters
to achieve a given degree of fit increases rapidly with dimensionality.
In this way, the simple scaling of volumes with radius in k
dimensions translates into a complexity penalty in the MDL and Bayesian
model selection methods. [The constant term (k/2)ln(2π)
has its origins in the details of the analysis and is less easy to
explain intuitively.]
A useful insight into the meaning of MDL is obtained by rewriting the criterion as:
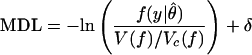 |
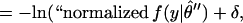 |
where δ ≡ ½ln(det J(θ̂)/det I(θ̂)) becomes negligible as N grows large when the truth lies within the family f. This rewriting provides a clearer picture of what MDL does in model selection. It selects the model with the highest value of the maximized likelihood per the relative ratio of distinguishable distributions [V(f)/Vc(f)]. We might call this the “normalized maximized likelihood.”
Generalizability, MDL, and Geometry.
Classic results in information theory confirm the efficacy of MDL and Bayesian methods in selecting models that predict well the statistics of later events in a sequence of data. The Bayesian perspective described above shows that the model preferred by Bayesian methods is the most likely truth in the sense of probability theory (also see, e.g., Theorem 1 in ref. 26). Concerning MDL, as the sample size increases, MDL is guaranteed to eventually pick a model that is optimal in the sense of minimizing the coding-theoretic description length of data drawn from the truth (see Theorem 1 in ref. 16). The ability to optimally encode long sequences of data can be interpreted in terms of generalizability, as forcefully argued by Rissanen in a classic paper (2).‡‡ Li and Vitányi (13) extensively discuss the relation between short encoding and generalizability in Chapter 5 of their book, albeit in an idealized setting. Barron and Cover's results imply that MDL and Bayesian methods are asymptotically consistent: given enough data, they will converge to the truth if it is present within the model families (1). (With a little work, our setup may be translated into theirs.) This also implies that the (suitably defined) generalization error of models selected by MDL and Bayesian methods decreases as the sample size increases (see Proposition 4 of ref. 1) when the truth lies in the model class. A related theorem, where the generalization error is defined as the expected mean squared prediction error made by the model inferred from the data (with the expectation taken in the true distribution) is provided by Li and Vitány (13) (Theorem 5.2.1). However, their precise setup is sufficiently different from ours that this particular theorem can only suggest, not prove, that practical MDL methods generalize well. The quest to prove similar strong theorems when the truth is not a member of the model class and when the sample size is small†† is a topic of active research by statisticians and information theorists. It is expected that MDL and Bayesian methods will continue to perform well even in these cases, because these information theoretic criteria select models that contain a large relative volume of distinguishable distributions lying close to the truth. One of these distributions should be selected when fitting a model to a sample. A second sample should also define an empirical distribution close to the truth and therefore should be described well by the model§§, i.e., the model generalizes well. A large Vc/V implies that a large fraction of the distributions indexed by the model will generalize well in this manner. This suggests that statistical fluctuations in the data leading to different choices of best-fitting model parameters will have less effect on generalization performance for models with large Vc/V.
A geometric perspective has taught us that the size of a model manifold in the space of distributions is what matters in measuring a model's complexity, not the apparent complications of its functional form or the number of parameters. The latter two properties of a model can be red herrings, as they are simply the apparatus by which a collection of distributions defined by the model is indexed. When examined individually, they can lead to an insufficient, even misleading, understanding of complexity. Neither the parameterization nor the specific functional form used in indexing is relevant, so long as the same collection of distributions is catalogued on the manifold. For example, the following two models, though assuming different functional forms, are equivalent and equally complex in the geometric sense: Model A, y = (ab/(ab + (1 − a)(1 − b))) + error, (0 < a, b < 1), and Model B, y = (1/(1 + eα+β)) + error (−∞ < α, β < ∞), where the error has zero mean and follows the same distribution for both models. Here, the parameters θ = (a, b) of Model A are related to the parameters η = (α, β) of Model B through α = ln((1 − a)/a) and β = ln((1 − b)/b)).
Example Application
Geometric complexity and MDL are a powerful pair of model evaluation tools. Used together, they elicit a deeper understanding of the relationship between models, as the following example shows. Consider two models from psychophysics describing the relationship between physical dimensions (e.g., light intensity) and their psychological counterparts (e.g., brightness): y = axb + error (Stevens' model) and y = a ln(x + b) + error (Fechner's model). We generated data samples from each model and then fitted both models to each data set. Under AIC or BIC, Stevens' model was always selected more often than Fechner's model, even when the data were generated by the latter (63% vs. 37%; Table 2). That is, AIC and BIC overestimated the generalizability of Stevens' model relative to Fechner's model, suggesting that the former is more complex than the latter. Implicit in the argument is the claim that Stevens' model fitted Fechner's data best because the former has more distinguishable distributions at its disposal, most of which enabled it to capture random noise rather than the underlying regularity. Calculation of the geometric complexity of each model confirms this suspicion, as Townsend speculated 25 years ago (27). Stevens' model is more complex than Fechner's, with the complexity difference being equal to 3.804. Given the logarithmic relationship between geometric complexity and the number of distinguishable distributions, this means that for every distribution for which Fechner's model can account, Stevens' model can describe about e3.804 ≈ 45 distributions. Obviously, this complexity difference between the two models must be due to the functional form, because they have the same number of parameters. When MDL was used, the model recovery rate was nearly perfect for both models, because the effect of complexity due to functional form was properly incorporated. As this example shows, accounting for the complexity of models is essential for a well-founded model selection procedure.
Table 2.
Model recovery rates of two psychophysics models
| Selection method | Model fitted | Data from Stevens | Data from Fechner |
|---|---|---|---|
| AIC, BIC | Stevens | 100% | 63% |
| Fechner | 0% | 37% | |
| MDL | Stevens | 99% | 2% |
| Fechner | 1% | 98% |
The table shows the percentage of samples in which the particular model was selected. One thousand samples were generated from each model using the same four points for x, which ranged from 1 to 4 in increments of 1. The random error was normally distributed with a mean of zero and a standard deviation of 1. The parameters values used to generate the simulated data were a = 2, b = 2 for Stevens' model and a = 2, b = 5 for Fechner's model. In computing the geometric complexity as well as in estimating best-fit values of the parameters, the following parameter ranges were assumed: 0 < a < ∞, 0 < b < 3 for Stevens' model, and 0 < a, b < ∞ for Fechner's model.
Summary and Conclusion
Model selection can proceed confidently when a well-justified and intuitive framework for its central concept, complexity, is available. We have shown that the geometry of the space of probability distributions provides such a framework. Rather than including seemingly disparate measures of complexity such as the number of parameters and the functional form, we construct the geometric complexity of a model by counting the number of distributions it indexes. This quantity is manifestly invariant under reparametrizations and is a basic ingredient for assessing the complexity of a statistical model in the MDL and Bayesian selection methods. These tools provide powerful methods of evaluating the effectiveness of models and the relationships between them.
Acknowledgments
The authors thank W. Batchelder, D. Bamber, M. Browne, P. Grunwald, J. Rissanen, and J. Townsend for valuable comments, and Shaobo Zhang for his assistance with the analysis reported in Table 2. We are especially grateful to D. Bamber, whose careful reviews strengthened this paper, W. Batchelder, who provided our example of two equivalent models with differing model equations (models A and B), and Peter Grunwald for his close reading of this text and suggestions for improvement. I.J.M. and M.A.P. were supported by National Institute of Mental Health Grant MH57472. V.B. was supported by the Society of Fellows and the Milton Fund of Harvard University, and by National Science Foundation grant NSF-PHY-9802709.
Abbreviations
- MDL
minimum description length
- MSE
mean squared error
- AIC
Akaike Information Criterion
- BIC
Bayesian Information Criterion
- SC
stochastic complexity
Appendix
Suppose θp and θq index two distributions in the family f and that y is a sample of size N drawn from one of θp or θq. In the model selection context, we test distinguishability of these distributions by asking how well we can guess which one produced y. Let αN and βN be the probabilities that θp is mistaken for θq and vice versa. There is a bound on how small these error probabilities can be made. Suppose we require that αN ≤ ɛ. Then, for large N, Stein's lemma says that the other error probability βN will exceed any given β* in an ellipsoid defined around θp, where κ ≡ −ln(β*) + ln(1 − ɛ) ≥ (N/2)∫i,j=1kIijδθiδθj (14). (Here Iij is the Fisher Information defined earlier.) If β* is large, the distributions within this region are not very distinguishable and should not be counted as separate distributions while working with N data points. See refs. 10, 14, and 17 for further technical details.
To implement this procedure, imagine partitioning the manifold into many (M) small cubes, where the cube centered on θj has a coordinate volume dUj = Πi=1k ≡ dθi ≡ dθ. For very large N, each such cube contains many ellipsoids of indistinguishability, as defined in the text. The Fisher Information will be basically constant within such tiny regions, and so the ellipsoids have a volume
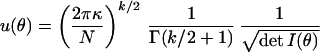 |
at any fixed N and κ. The number of distinguishable distributions within dUj is dUj/u(θj).
The ratio of distinguishable distributions within the cube at θj to those contained in the entire family is
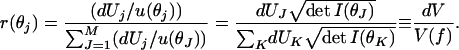 |
Happily, the dependence of the ellipsoid volumes on the parameters N and κ has cancelled out. So we can take the limit N → ∞, followed by M → ∞ to recover the continuum. We are left with
 |
This is the ratio of the volume of the infinitesimal region
dθ to the volume of the parameter manifold as a whole. So,
for the purpose of determining such ratios, the appropriate volume
integration measure on a model manifold is dV =
dθ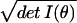 . The procedure of first
turning the manifold into a lattice and then removing the
discretization has “divided out” the volume occupied by
indistinguishable distributions.
. The procedure of first
turning the manifold into a lattice and then removing the
discretization has “divided out” the volume occupied by
indistinguishable distributions.
Footnotes
In situations where the mean h or the distribution on ei is not well defined, we simply model the full conditional probability Pr(Y|X).
Different error measures including MSE can be appropriate in different problems. In a statistical context, the logarithmic loss function Err(Data|Model) = − ln Pr(Data|Model) is attractive because it directly measures the ability of the inferred model to predict the true data statistics.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170283897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170283897
These guarantee that L(f) can be interpreted as the number of bits required to describe f using some code.
Some technical conditions are required; see, e.g., ref. 25.
Rissanen provides a theorem (Theorem 1) that strongly suggests that code-length-minimizing models should also generalize well. He also formally shows that, under certain conditions, code-length-minimizing models provably generalize well, though the notion of generalizability he uses is somewhat different from that used here.
Any given learning strategy may perform poorly on very small data samples taken from specifically constructed adversarial examples. When sample sizes are tiny (e.g., one data point), analytic model selection criteria can behave counterintuitively. However, this is not surprising: very little can be inferred from scanty data.
There are triangle inequalities governing natural distances (e.g., relative entropy, L2 norm) between distributions. These guarantee that two distributions close to a third are also mutually close; see ref. 14.
References
- 1.Barron A R, Cover T M. IEEE Trans Inf Theory. 1991;37:1034–1054. [Google Scholar]
- 2.Rissanen J. IEEE Trans Inf Theory. 1984;30:629–636. [Google Scholar]
- 3.Myung I J. J Math Psychol. 2000;44:190–204. doi: 10.1006/jmps.1999.1283. [DOI] [PubMed] [Google Scholar]
- 4.Linhart H, Zucchini W. Model Selection. New York: Wiley; 1986. [Google Scholar]
- 5.Akaike H. In: Second International Symposium on Information Theory. Petrox B N, Caski F, editors. Hungary: Academi Kiado; 1973. pp. 267–281. [Google Scholar]
- 6.Schwartz G. Ann Stat. 1978;6:461–464. [Google Scholar]
- 7.Rissanen J. Ann Stat. 1986;14:1080–1100. [Google Scholar]
- 8.Kolmogorov A N. IEEE Trans Inf Theory. 1968;14:662–664. [Google Scholar]
- 9.Rissanen J. Econometric Rev. 1987;6:85–102. [Google Scholar]
- 10.Balasubramanian V. A Geometric Formulation of Occam's Razor for Inference of Parametric Distributions, Princeton physics preprint PUPT-1588. NJ: Princeton; 1996. [Google Scholar]
- 11.Chaitin G J. J Assoc Comp Machn. 1966;13:547–569. [Google Scholar]
- 12.Solomonoff R J. Inform Contr. 1964;71:224–254. [Google Scholar]
- 13.Li M, Vitányi P. An Introduction to Kolmogorov Complexity and Its Applications. New York: Springer; 1997. [Google Scholar]
- 14.Cover T, Thomas J. Elements of Information Theory. New York: Wiley; 1991. [Google Scholar]
- 15.Grunwald P. J Math Psychol. 2000;44:133–152. doi: 10.1006/jmps.1999.1280. [DOI] [PubMed] [Google Scholar]
- 16.Rissanen J. IEEE Trans Inf Theory. 1996;42:40–47. [Google Scholar]
- 17.Balasubramanian V. Neural Computation. 1997;9:349–368. [Google Scholar]
- 18.Amari S I. Differential Geometrical Methods in Statistics. Berlin: Springer; 1985. [Google Scholar]
- 19.Amari S I, Barndorff-Nielsen O E, Kass R E, Lauritzen S L, Rao C R. Differential Geometry in Statistical Inference. Hayward, CA: Institute of Mathematical Statistics; 1987. [Google Scholar]
- 20.Rao C R. Bull Calcutta Math Soc. 1945;37:81–91. [Google Scholar]
- 21.Efron B. Ann Stat. 1975;3:1189–1242. [Google Scholar]
- 22.Amari S I. Electr Comm Jap. 1983;66A:1–10. [Google Scholar]
- 23.Atkinson C, Mitchell A F. Sankhya: Indian J Stat. 1981;43:345–365. [Google Scholar]
- 24.Murray M K, Rice J W. Differential Geometry and Statistics. London: Chapman & Hall; 1993. [Google Scholar]
- 25.Clarke B S, Barron A R. IEEE Trans Inf Theory. 1990;36:453–471. [Google Scholar]
- 26.Vitanyi P, Li M. IEEE Trans Inf Theory. 2000;46:446–464. [Google Scholar]
- 27.Townsend J T. In: Philosophical Aspects of the Mind-Body Problem. Cheng C, editor. Honolulu: Honolulu Univ. Press; 1975. pp. 200–218. [Google Scholar]


