Abstract
Ten structural genes from the Capsicum (pepper) carotenoid biosynthetic pathway have been localized on a (Capsicum annuum × Capsicum chinense)F2 genetic map anchored in Lycopersicon (tomato). The positions of these genes were compared with positions of the same genes in tomato when known, and with loci from pepper, potato, and tomato that affect carotenoid levels in different tissues. C2, one of three phenotypically defined loci determining pepper fruit color, cosegregated with phytoene synthase. The capsanthin–capsorubin synthase (Ccs) locus, shown previously to cosegregate with Y, another pepper fruit color locus, mapped to pepper chromosome 6. Other structural genes in pepper corresponded to loci affecting carotenoid production as follows: Ccs in pepper and the B locus for hyperaccumulation of β-carotene in tomato fruit mapped to homeologous regions; the position of the lycopene β-cyclase gene in pepper may correspond to the lutescent-2 mutation in tomato; and the lycopene ɛ-cyclase locus in pepper corresponded to the lycopene ɛ-cyclase locus/Del mutation for hyperaccumulation of δ-carotene in tomato fruit. Additional associations were seen between the structural genes and previously mapped loci controlling quantitative variation in pepper and tomato fruit color. These results demonstrate that comparative analyses using candidate genes may be used to link specific metabolic phenotypes and loci that affect these phenotypes in related species.
Studies of biosynthetic pathways in plants have linked biochemical variability to cloned structural and regulatory genes. In maize, the only case to date where the complete set of genes from a pathway has been examined, the maysin content of corn silks was associated with alleles of both regulatory and structural loci (1–4). Other results from both maize (5, 6) and Arabidopsis thaliana (7) have shown that variability in quantitative measurements of overall enzymatic activity can be attributed to structural loci or closely linked elements. If these studies represent a general trend, the power of this approach will be increased by comparative analyses where homologous genes affect similar phenotypes in different plant species. Although higher plants produce a diverse array of secondary metabolites often unique for a particular taxon, the conserved genetic basis of similar biosynthetic products is implied by comparative studies localizing quantitative trait loci (QTL) to homeologous parts of related genomes (e.g., refs. 8–10) and the tagging/cloning of two QTL controlling erucic acid production in Brassica napus by using candidate structural genes cloned from Arabidopsis (11).
Carotenoids are the red, orange, and yellow molecules that act as photoprotective agents and accessory light-harvesting pigments, and add nutritional and ornamental value to plants (reviewed in refs. 12–14). Because of their importance, the enzymes in this pathway have been characterized and the structural genes have been cloned from a wide array of higher plants (refs. 13 and 15–17 and references therein), including pepper (Capsicum annuum L.) (18–27) and tomato (Lycopersicon esculentum L.) (28–30). Further, the genetic basis of variation in fruit color due to alterations in carotenoid content has been well established in tomato and, to a lesser extent, pepper. Mature tomato fruits exhibit a wide range of colors, and many mutations (see ref. 31 and references therein), and QTL (32, 33) affecting fruit color have been characterized and mapped. In pepper, interactions between three unlinked genes are postulated to produce eight color classes, ranging from red to white (34); however, the identity of genes responsible for this fruit color variability in the Solanaceae remains unknown except in three cases. Two fruit color mutants in tomato, Del (red to orange) and r (red to yellow), and one mutant in pepper, Y (red to yellow), control qualitative fruit color shifts and have been shown to be identical or tightly linked to structural genes in the carotenoid pathway (30, 35–37).
To date, no study has systematically examined the relationship between structural genes in a biosynthetic pathway and related phenotypic variability by using a comparative genetic system. If the candidate gene approach to these traits can be extended by using comparative genetic maps, nonoverlapping sets of mutants assembled across plant species may prove more informative about the function and evolution of that locus than the allelic variability present in one species. Further, candidate genes for phenotypically defined quantitative and qualitative loci may be immediately clear for the array of species. The objectives of this study, therefore, were to (i) map the cloned structural genes of the carotenoid biosynthetic pathway in the Solanaceae and (ii) examine the extent to which structural genes or tightly linked sequences affect inherited qualitative and quantitative phenotypic variability in a set of plant species linked by a comparative genetic map.
Materials and Methods
Genetic Mapping of Carotenoid Biosynthetic Enzyme Loci.
Map positions for the carotenoid biosynthetic enzymes were obtained by using the previously described comparative genetic linkage map of pepper (38). The map was constructed by using 75 (Capsicum annuum cv. NuMex RNaky × Capsicum chinense PI 159234)F2 plants genotyped with amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), and tomato- and pepper-derived genomic and cDNA probes used in restriction fragment length polymorphism (RFLP).
PCR primers were designed by using GenBank sequences (Table 1) for geranylgeranyl pyrophosphate synthase (Ggpps); phytoene synthase (Psy); phytoene desaturase (Pds); ζ-carotene desaturase (Zds); and zeaxanthin epoxidase (Ze). Primers used for capsanthin capsorubin synthase (Ccs) were from ref. 36. Genomic DNA from C. annuum NuMex RNaky was used for the amplification of Psy, Zds, Ze, and Ccs. Total mRNA was isolated from C. annuum NuMex RNaky by the method of Dunn (39). Ggpps was amplified by reverse transcription (RT)-PCR according to ref. 40, using 2 μl of cDNA, 0.8 mM dATP, dCTP, dGTP, dTTP, and forward and reverse primers, and the standard concentration of the supplier's PCR buffer with Mg2+ and 1.25 units of Taq DNA polymerase (Roche Molecular Biochemicals, Indianapolis, IN) in a 25-μl reaction volume. Pds was likewise amplified by RT-PCR and cloned in pGEMT (Promega, Madison, WI). The cloned tomato genes, lycopene β-cyclase (CrtL-b), lycopene ɛ-cyclase (CrtL-e), and β-carotene hydroxylases (CrtZ-1, CrtZ-2), were obtained from J. Hirschberg (Hebrew University of Jerusalem).
Table 1.
Primer sequences used for amplification of carotenoid genes from pepper
| Gene | Genbank accession no. | Fragment size, bp | Primer sequence (5′–3′) |
|---|---|---|---|
| Ggpps | X80267 | 1,200 | F GAACCTTGTTGATTTATGGGC |
| R CCAACATAAGCACACTGAAAG | |||
| Psy | X68017 | 1,200 | F TGGGCATCGCACCTGAATCAA |
| R GTCCAGTATCCTGCGGTACAA | |||
| Pds | X68058 | 420 | F TTCGACTTGTTTCTGCTGTCA |
| R CATCCCTTGCCTCCAGCAGTA | |||
| Zds | X89897 | 400 | F GCTCCAAAAGGGCTATTTCC |
| R TCCCATTTCAATGTGGTTCC | |||
| Ze | X91491 | 1,900 | F ATGGCATAAGGTCTAAGGTAC |
| R CTCAGATAGTCTGCAATGTTG | |||
| Ccs | X77289 | 1,490 | F CTAATGGAAACCCTTCTAAAGC |
| R AATTCAAAGGCTCTCTATTGCT |
F, forward; R, reverse.
The genes were amplified for use as RFLP probes according to one of two PCR profiles. Reaction conditions for Pds, Zds, CrtL-b, CrtL-e, CrtZ-1, and CrtZ-2 consisted of 25.0 ng of plasmid DNA, 0.8 mM dATP, dCTP, dGTP, and dTTP, 0.1 mM forward and reverse primers, the standard concentration of buffer, and 2.5 units of Taq polymerase in a total volume of 100 μl. The conditions for amplifying Ccs, Psy, and Ze were 15 ng of genomic DNA, 0.4 mM dATP, dCTP, dGTP, and dTTP, 0.1 mM forward and reverse primers, the standard concentration of buffer, and 1.25 units of Taq polymerase in a total volume of 25 μl. The RT-PCR amplification product was used for Ggpps. PCR products were then used as probes in RFLP and mapped as described (38).
Tests for Associations Between C1, C2, and the Carotenoid Biosynthetic Enzymes.
A population segregating for mature fruit color was created by crossing C. chinense PI 152225 (red) and C. annuum 4751 (white) (36). A total of 174 (C. annuum 4751 × C. chinense PI 152225) × C. annuum 4751 BC1F1 individuals were grown in the greenhouse at Bet Dagan, Israel. Five to 10 fruits were selected from each plant, stored for 1 week, then grouped according to color.
Associations between C1 and C2 and the carotenoid genes were examined by using the cloned carotenoid genes as probes in RFLP. DNA was extracted from leaves of parents and BC1F1 individuals, and the genes were then used to probe filters containing DNA from 12 red-fruited and 12 white-fruited BC1F1 individuals. Genes showing RFLP segregating with one color group were then used to genotype the entire BC1F1 population.
Tagging the Y, C1, and C2 Loci in Pepper.
Anonymous DNA sequences were used to tag the pepper color loci by bulked segregant analysis (41). DNA bulks consisting of 11 red- and 11 white-fruited BC1 progeny were screened with 500 primers from Operon Technologies (Alameda, CA; kits K–O, Q–U), Advanced Biotechnologies (Epsom, Surrey, U.K.; kits 1–5), and the University of British Columbia (primers 1–200) in RAPD analyses as previously described (42). Primers with RAPD bands only in the red-fruit bulk were tested against another set of four different color bulks, and primers displaying diagnostic patterns were then used to genotype all BC1F1 individuals.
Results and Discussion
Associations Between Classical Color and Carotenoid Structural Genes.
Color classification within the backcross population.
The predicted range of fruit color from red to white was present in the BC1F1 population (ref. 34; Table 2); however, continuous variation in fruit color made delimiting classes difficult. Consequently, some color classes were left joined, defining four color groups—red (red to light red), peach (salmon pink to peach), orange (orange to orange-yellow), and cream (lemon yellow to white)—used for the remainder of the experiment (Table 2). The division into four color groups indicated that two major genes were segregating in the population, and the data fit the expected Mendelian ratio for unlinked loci (1:1:1:1) with χ2 = 2.96, 3 df, P = 0.397. The red- and white-fruited individuals used for the testing and tagging were taken from the extremes of their respective color class.
Table 2.
Color and proposed genotypes for fruit color classes
| This
study
|
Hurtado-Hernandez and Smith
|
||||||
|---|---|---|---|---|---|---|---|
| Genotype
|
Fruit color | Genotype | Fruit color | ||||
| Ccs | Psy | ||||||
| Y+ | C1+ | C2+ | Red | Y+ | C1+ | C2+ | Red |
| Y+ | c1 | C2+ | Red | Y+ | c1 | C2+ | Light red |
| Y+ | C1+ | c2 | Peach | Y+ | C1+ | c2 | Orange |
| Y+ | c1 | c2 | Peach | Y+ | c1 | c2 | Pale orange |
| y | C1+ | C2+ | Orange | y | C1+ | C2+ | Orange-yellow |
| y | c1 | C2+ | Orange | y | C1+ | c2 | Pale orange-yellow |
| y | C1+ | c2 | Cream | y | c1 | C2+ | Lemon-yellow |
| y | c1 | c2 | Cream | y | c1 | c2 | White |
The classes were observed in a segregating population derived from a cross between a red-fruited parent, C. chinense PI 152225, and a white-fruited parent, C. annuum 4751, compared with genotypic classes predicted by the Hurtado-Hernandez and Smith genetic model (34).
Testing associations between classical color and carotenoid structural genes.
The screen of the ten carotenoid genes against the red and white BC1F1 individuals showed that other than Ccs (36), only Psy exhibited a polymorphism segregating with red color. When the entire BC1F1 population was probed with Psy, the C. chinense allele was present in the red and orange color groups and absent in the peach and cream groups, consistent with the action attributed to the C2 gene (34) (Table 2). Results for Ccs confirmed that the red and peach groups were Y+, whereas the orange and cream groups were y.
The differences between the red vs. peach, and the orange vs. cream groups appeared to be due to different levels of the same pigments, suggesting that Psy/C2 may be a major gene acting as an overall regulator of carotenoid amounts. HPLC analyses showing that peach and cream classes contain low levels of carotenoids (0.065 mg/g dry weight and 0.043 mg/g dry weight, respectively) compared with red fruits (2.6–5.6 mg/g dry weight) (43) support this putative role of Psy/C2 as a rate-limiting factor in carotenoid production. Further support for this hypothesis comes from studies in tomato, where lines over-expressing an antisense Psy showed a reduction in fruit carotenoid levels (44). The difference(s) between alleles could be in either cis-acting regulatory sequences or the structure of the enzyme; both types of polymorphism have been seen in studies relating QTL and structural genes (e.g., refs. 7 and 11).
Tagging pepper color loci by using RAPDs.
Eight primers produced polymorphisms between the red and white bulks. These primers were then used to screen bulks of 10 BC1 plants each made from the four color groups. Primers OP L-09, OP N-7, UBC-19, UBC-131, and UBC-182 amplified fragments in the red and peach bulks that were absent in the cream and orange bulks, suggesting linkage in cis to Y+ (Table 2). From these primers, OP N-7 was chosen to screen the entire BC1 population, and a 500-bp band showed linkage with Ccs/Y at a distance of 7 centimorgans (cM).
Primers OP O-12, OP S-15, and UBC-183 amplified fragments in the red and orange bulks, whereas the bands were absent from the peach and cream bulks, indicating linkage in cis to C2+ (Table 2). Primers OP O-12 and UBC-183 were used to screen the whole BC1 population, and amplified fragments (700 and 900 bp, respectively) were found to be 1 cM apart from each other and 36 cM away from Psy/C2.
Comparison with the Hurtado-Hernandez and Smith model.
A summary of the data is presented in Table 2, along with the classical model of Hurtado-Hernandez and Smith (34). As reported previously, the Y gene is Ccs (35, 36). The segregation data for Psy suggest that it is the gene previously designated C2, which has a significant effect on the total level of carotenoid accumulation in fruit. The effects of these two genes, Y and C2, on fruit carotenoid content were confirmed by HPLC results showing that fruits in the peach class do contain capsanthin and capsorubin, the predominant pigments of red peppers, but at a level approximately 1/100 that of red fruits (43). No reference genotype is available for C2, but the locus segregating with Psy matches the description of this factor by Hurtado-Hernandez and Smith (34). Therefore we conclude that Psy is the candidate for C2.
The C1 gene also apparently affects the amount, rather than the type, of carotenoids present. The range of color observed both across the entire BC1 population and within the four color groups indicates a quantitative component of fruit color. Our inability to detect C1 in this population may be due to either absence or homozygosity of an allele that was segregating in the population analyzed by Hurtado-Hernandez and Smith. Future evaluations using analytical measures of extractable carotenoids should clarify the nature and genomic locations of QTL affecting pepper fruit color.
Complicating this issue, however, are data indicating that pepper fruit color is subject to background effects (I.P., unpublished data) and the possibility that similar fruit colors may be caused by different combinations of pigments (45). In fact, the genotype of orange-colored varieties may be either y C2+ or Y+ c2, depending on the source of the germ plasm (36), such that orange peppers may or may not contain capsanthin and capsorubin (43, 46). The changes suggested in Table 2 conform with our data and the crosses made by Hurtado-Hernandez and Smith; however, they are made on the basis of the alleles tested and may require further refinement from results of other crosses.
Mapping the Structural Genes in the Carotenoid Biosynthetic Pathway.
Segregating polymorphisms defined map positions in pepper for all 10 structural genes. The previous alignment of this pepper map with the tomato and potato maps (38) allowed comparisons between the positions of these loci and phenotypically defined genes and QTL affecting carotenoid content (ref. 31 and references therein; refs. 32–34, 47). These comparisons are based on the assumption of conservation of both macro- and microcolinearity, and therefore are subject to the limitations of comparative mapping. In at least one case (Psy), the colinearity of the segments containing homologous genes is interrupted by a translocation. New associations suggested by results of this study (Fig. 1) define hypotheses of relationship between a particular candidate gene and the character in question that await rigorous tests of relationship in appropriate populations. The results for each structural gene, in order of appearance of gene products in the biosynthetic pathway, are presented below.
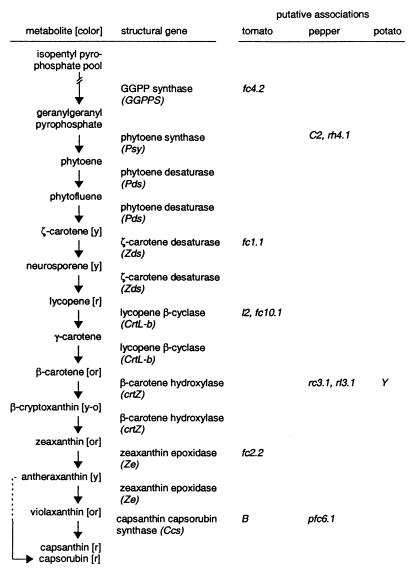
Figure 1.
Simplified version of the carotenoid biosynthetic pathway in pepper fruit and putative correspondences with quantitative and qualitative organ color loci identified in the Solanaceae revealed by this study. The metabolic intermediates of the carotenoid pathway and structural genes responsible for their catalysis are shown, with the exception of the conversion of lycopene to δ-carotene by lycopene ɛ-cyclase. Putative associations between these genes and previously defined qualitative (Xa, l2, B) and quantitative (fc1.1, fc2.2, fc4.2, fc10.1) loci in tomato, potato (Y), and pepper (pfc6.1, rc3.1, rl3.1, rh4.1), suggested by data from this study and shown in Fig. 2 are summarized. Previously defined identities (phytoene synthase–yellow-flesh and lycopene ɛ-cyclase–Del) in tomato and correspondences in pepper (capsanthin capsorubin synthase–Y) are not shown. The color of the intermediate is indicated in brackets: [y] = yellow, [or] = orange, [y-o] = yellow-orange, and [r] = red.
Geranylgeranyl pyrophosphate synthase.
Ggpps mapped to pepper chromosome 4, between markers CD62a/CD39b and CT253a (Fig. 2). Survey results indicated a single copy in the pepper genome. In tomato, a QTL (fc4.2) affecting a small proportion (R2 = 0.05) of mature red fruit color intensity was mapped in this region of chromosome 4 (32) (Fig. 2).
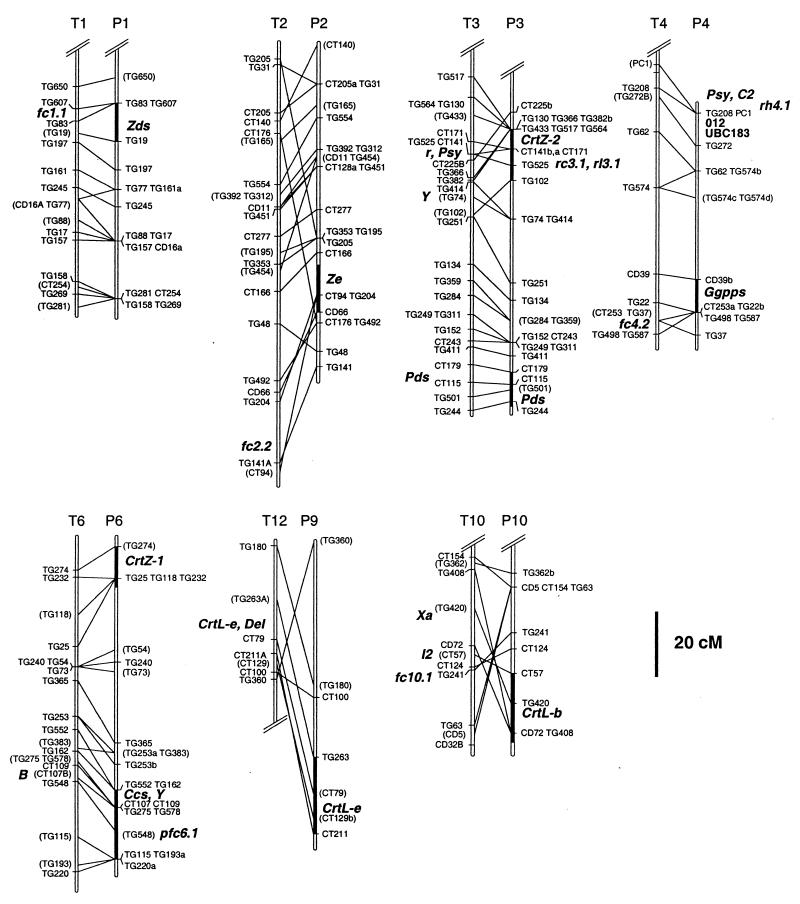
Figure 2.
Comparative map of pepper and tomato chromosomes, showing the position of carotenoid structural genes and phenotypically defined loci. T1, T2, T3, T4, T6, T10, and T12 refer to tomato chromosomes from ref. 57. P1, P2, P3, P4, P6, P9, and P10 refer to pepper chromosomes defined in ref. 38. Double slash marks at termini indicate that partial chromosomes are shown. Markers consist of tomato genomic (TG) and tomato cDNA (CD and CT) clones. Maps were aligned according to ref. 38, and lines between chromosomes connect presumed orthologues. Pepper genes are geranylgeranyl pyrophosphate synthase (Ggpps), phytoene synthase (Psy), phytoene desaturase (Pds), ζ-carotene desaturase (Zds), lycopene β-cyclase (CrtL-b), lycopene ɛ-cyclase (CrtL-e), β-carotene hydroxylase-1 and -2 (CrtZ-1 and -2), zeaxanthin epoxidase (Ze), and capsanthin–capsorubin synthase (Ccs). The darkened chromosomal segment associated with each of the carotenoid genes indicates the support interval for each gene, including all positions with ΔLOD ≤ 2. O-12 and UBC183 are RAPDs linked to C2. Pepper QTL are pfc6.1, rc3.1, rl3.1, and rh4.1 (50, 55). Tomato genes are Psy, Pds, and CrtL-e. Tomato mutants are lutescent-2 (l2), Xa, Delta (Del), yellow-flesh (r), and B. Tomato map positions are from refs. 30, 31, 56, and 57. Tomato QTL (fc2.2, fc4.2, fc10.1) are from ref. 32 and fc1.1 is from ref. 33. The Y locus of potato shown on T3 was mapped in ref. 47. Placement of the locus designation reflects the best inferred position for the locus on this map.
Phytoene synthase.
As previously reported for tomato (48, 49) and pepper (25), at least two Psy homologues were detected, but only one was segregating in this population. The segregating homologue mapped to the end of chromosome 4 approximately 20 cM from TG507 (Fig. 2) and initially did not appear to correspond to either of the Psy positions mapped on tomato chromosomes 2 and 3 (49). Another marker tightly linked to a Psy homologue on tomato chromosome 3 (TG621), however, is also found at the top of pepper chromosome 4 (38), indicating a small translocation. The two RAPD fragments (O-12 and UBC183) linked to C2 also map to this region of chromosome 4 (Fig. 2). In pepper, a QTL affecting the red hue of fruit (rh4.1, R2 = 0.12) was recently mapped to this region (50) (Fig. 2). The position of this QTL suggests that it may be the result of qualitative differences in the structure or regulation of Psy in this population, implicating Psy as a source of both qualitative and quantitative variability in carotenoid biosynthesis.
Phytoene desaturase.
A single copy of Pds was identified in pepper and mapped to pepper chromosome 3, corresponding to the position reported for tomato (51) (Fig. 2). No organ color loci or QTL have been identified in the homeologous regions of pepper, potato, or tomato.
ζ-Carotene desaturase.
Zds was mapped to pepper chromosome 1 in a region homeologous to tomato chromosome 1. A second band was detected but was not polymorphic in this population. The position of Zds corresponded with a minor (R2 = 0.03) QTL (fc1.1) for fruit color intensity mapped in tomato (33) (Fig. 2).
Lycopene ɛ-cyclase.
A single copy of CrtL-e was detected in pepper and mapped to chromosome 9 between CT211 and TG623, with the most likely position 6 cM proximal to CT211 (Fig. 2). This position corresponds to the CrtL-e locus on tomato chromosome 12 (30). In tomato, an up-regulated allele of CrtL-e causes the Del mutation, resulting in fruit that ripen to orange as a consequence of hyperaccumulation of δ-carotene instead of lycopene (30).
Lycopene β-cyclase.
Two homologues of CrtL-b were detected in pepper. One mapped to chromosome 10 between CD72 and A255 (Fig. 2). Rearrangements in this region between pepper and tomato make precise comparisons difficult, but at least one, and possibly two, tomato carotenoid biosynthetic mutants map to this region (Fig. 2). One mutation is lutescent-2 (l2) which produces numerous pleiotropic effects, including premature yellowing of leaves because of photobleaching, delayed onset of red pigment development in fruit, and reduced levels of β-carotene and xanthophylls (52). CrtL-b is the enzyme that converts lycopene to β-carotene and δ-carotene to ɛ-carotene; therefore, a reduction in the activity of CrtL-b could reduce the amounts of β-carotene, xanthophylls, and other downstream carotenoids. Although CrtL-b has been cloned from tomato (29), no map position has been reported. These data suggest that the lutescent-2 mutant could be due to either a structural or a cis-acting regulatory mutation in CrtL-b.
The Xa mutation of tomato also maps to chromosome 10 (31). Although this mutation is not reported to be the result of a disruption in carotenoid biosynthetic activity, many of the effects of this locus are similar to those of other known carotenoid biosynthetic mutants. Homozygous Xa plants are inviable, a phenomenon commonly seen in maize carotenoid mutants (53). Xa mutants are also dwarfed and, similar to l2 plants, the leaves of Xa plants are yellow. Either l2 or Xa could be a consequence of altered expression or function of CrtL-b. In addition to these qualitative mutations, a QTL (fc10.1, R2 = 0.06) affecting fruit color intensity has been identified in this region of tomato (32) (Fig. 2). We were unable to assign a position in our population for the second pepper CrtL-b homologue.
β-Carotene hydroxylases.
Both CrtZ-1 and CrtZ-2 gave identical polymorphisms on the (C. annuum × C. chinense)F2 filters. Two copies were detected with both probes, as expected given the sequence similarity of the two genes in pepper (27) and tomato (J. Hirschberg, personal communication). One polymorphism was mapped to chromosome 3 and the other to chromosome 6 (Fig. 2). Determining the correspondence between genes and positions was not possible in the primary mapping population, but by using a separate C. annuum × C. chinense population it was determined on the basis of differences in band intensity that CrtZ-2 corresponded to the position on chromosome 3, whereas CrtZ-1 corresponded to the position on chromosome 6 (I.P., unpublished data).
Two QTL, one affecting red chroma (rc3.1, R2 = 0.13) and the other, red lightness (rl3.1, R2 = 0.16), were identified on chromosome 3 in an intraspecific C. annuum population (50) (Fig. 2). The tomato region homeologous to the CrtZ-2 position contains the yellow-flesh mutation, but it has previously been shown that the yellow-flesh mutant results from alterations in Psy (37). In potato, the Y locus has also been mapped to this region (47). Alleles at this locus cause tubers to turn from white to orange or yellow because of an accumulation of zeaxanthin (54). The Y locus cosegregated with TG74 in an earlier mapping study (47). While the position for CrtZ-2 in pepper does not include TG74, the region TG366–TG102 in pepper may be homeologous with the segment containing Y in potato (Fig. 2). CrtZ-1/2 encode the enzymes that convert β-carotene to zeaxanthin, which also suggests CrtZ-2 or a linked regulatory element as a candidate gene for Y in potato, although we cannot rule out Psy as a candidate in view of its position in tomato and the existence in pepper of an unmapped Psy homologue.
Zeaxanthin epoxidase.
Two Ze homologues were detected in pepper; one mapped to pepper chromosome 2 (Fig. 2), whereas the other was not polymorphic. An inversion on this chromosome differentiates pepper and tomato, but the position of Ze in pepper may correspond to a minor (R2 = 0.06) fruit color intensity QTL (fc2.2) of tomato (32).
Capsanthin capsorubin synthase.
Ccs, a single-copy gene in pepper, mapped to chromosome 6, 1 cM away from CT109 (Fig. 2). A QTL affecting the intensity of mature red color (pfc6.1) was detected in this region in the interspecific mapping population (55). This position also corresponds to the B locus in tomato (56) (Fig. 2), defined morphologically by the hyperaccumulation of β-carotene in fruits. Previous attempts in tomato to associate polymorphism in CrtL-b with the B locus were unsuccessful (29); however, the B gene has recently been cloned and shown to encode a novel lycopene β-cyclase (J. Hirschberg, personal communication; ref. 58). Ccs has also been reported to possess lycopene β-cyclase activity similar to CrtL-b (26, 58).
Conclusions
To our knowledge, this is the first study in which all of the structural genes from a pathway needed to produce agriculturally significant phenotypic variation in multiple genera have been placed on a comparative genetic map. At least one homologue of every enzyme examined except Pds appeared to correspond with loci previously reported to affect organ color in at least one of the three crop species. We found this level of correspondence surprising, given that the phenotypic data are derived from only a limited amount of germ plasm in each genus. In two cases, CrtZ-2 and Ccs, organ color loci responsible for both qualitative and quantitative shifts in carotenoid content in different plant genera appeared to overlap, suggesting the possibility that orthologous loci in potato and pepper, or tomato and pepper, are involved.
As these hypotheses of relationship are tested, causative variation could be discovered in either the cis-acting regulatory sequences or the structural genes themselves. We hypothesize that regulatory differences may be more likely in this pathway because of the indispensable nature of carotenoids: structural mutations that alter the function of these genes would most likely have negative fitness consequences. The data from tomato showing that not all QTL were expressed in all environments also support the hypothesis that many of these alleles are mainly due to regulatory differences.
Finally, there are other tomato mutants that affect fruit carotenoid content (e.g., ghost, tangerine, and apricot), and QTL in pepper and tomato fruit color QTL, some apparently common to both species (50), that do not overlap with the positions of any carotenoid structural genes. As structural genes and cis-acting regulatory sequences are excused as candidates for these loci, it becomes more likely that the remaining genes/QTL may be involved in regulation of this or related pathways, or may act more globally in fruit development. In all cases, comparison of the structure, function, and regulation of orthologous structural genes should shed light on divergence in key biosynthetic pathways.
Acknowledgments
We acknowledge L. Landry, J. Jantz, and G. Moriarty for their technical assistance. We thank J. Hirschberg, D. Zamir, and G. Ronen for clones and access to unpublished data. We thank M. McMullen, E. Wurtzel, R. Grube, M. Cadle, and L. Landry for critical reading and suggestions that improved the manuscript. T.A.T. was supported by an assistantship provided by Seminis, Inc., to M.J. K.D.L. was supported by a Department of Energy/National Science Foundation/U.S. Department of Agriculture grant to the Research Training Group in Molecular Mechanisms of Plant Processes and a gift from Marie Lavallard. Financial and material support was provided by the California Pepper Commission/California Pepper Improvement Foundation (M.J.), Kalsec, Inc. (M.J.), U.S. Department of Agriculture National Research Initiative Competitive Grants Program Award 94-37300-0333 (M.J.), Binational Agricultural Research and Development Award IS-2389-94 (I.P. and M.J.), and a Chief Scientist of the Israeli Ministry of Agriculture Grant (I.P.).
Abbreviations
- QTL
quantitative trait locus or loci
- RAPD
random amplified polymorphic DNA
- RFLP
restriction fragment length polymorphism
- Ggpps
geranylgeranyl pyrophosphate synthase
- Psy
phytoene synthase
- Pds
phytoene desaturase
- Zds
ζ-carotene desaturase
- CrtL-b
lycopene β-cyclase
- CrtL-e
lycopene ɛ-cyclase
- CrtZ-1
CrtZ-2, β-carotene hydroxylases
- Ze
zeaxanthin epoxidase
- Ccs
capsanthin capsorubin synthase
- cM
centimorgan
Note Added in Proof.
Ref. 58, published while this paper was in press, reports that the B locus in tomato is homologous to Ccs.
References
- 1.Byrne P F, McMullen M D, Snook M E, Musket T A, Theuri J M, Widstrom N W, Wiseman B R, Coe E H. Proc Natl Acad Sci USA. 1996;93:8820–8825. doi: 10.1073/pnas.93.17.8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne P F, McMullen M D, Wiseman B R, Snook M E, Musket T A, Theuri J M, Widstrom N W, Coe E H. Crop Sci. 1998;38:461–471. [Google Scholar]
- 3.Lee E A, Byrne P F, McMullen M D, Snook M E, Wiseman B R, Widstrom N W, Coe E H. Genetics. 1998;149:1997–2006. doi: 10.1093/genetics/149.4.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMullen M D, Byrne P F, Snook M E, Wiseman B R, Lee E A, Widstrom N W, Coe E H. Proc Natl Acad Sci USA. 1998;95:1996–2000. doi: 10.1073/pnas.95.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Causse M, Rocher J P, Henry A M, Charcosset A, Prioul J L, de Vienne D. Mol Breeding. 1995;1:259–272. [Google Scholar]
- 6.Prioul J-L, Pelleschi S, Séne M, Thévenot C, Causse M, de Vienne D, Leonardi A. J Exp Bot. 1999;50:1281–1288. [Google Scholar]
- 7.Mitchell-Olds T, Pedersen D. Genetics. 1998;149:739–747. doi: 10.1093/genetics/149.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutschler M A, Doerge R W, Liu S C, Kuai J P, Liedl B E, Shapiro J A. Theor Appl Genet. 1996;92:709–718. doi: 10.1007/BF00226093. [DOI] [PubMed] [Google Scholar]
- 9.Blauth S L, Churchill G A, Mutschler M A. Theor Appl Genet. 1998;96:458–467. doi: 10.1007/s001220050762. [DOI] [PubMed] [Google Scholar]
- 10.Bonierbale M W, Plaisted R L, Pineda O, Tanksley S D. Theor Appl Genet. 1994;87:973–987. doi: 10.1007/BF00225792. [DOI] [PubMed] [Google Scholar]
- 11.Fourmann M, Barret P, Renard M, Pelletier G, Delourme R, Brunel D. Theor Appl Genet. 1998;96:852–858. [Google Scholar]
- 12.Cogdell R J, Gardiner A T. Methods Enzymol. 1993;214:185–193. [Google Scholar]
- 13.Cunningham F X, Jr, Gantt E. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- 14.Demmig-Adams B, Gilmore A M, Adams W W. FASEB J. 1996;10:403–412. doi: 10.1096/fasebj.10.4.8647339. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong G A, Hearst J E. FASEB J. 1996;10:228–237. doi: 10.1096/fasebj.10.2.8641556. [DOI] [PubMed] [Google Scholar]
- 16.Bartley G E, Scolnik P A. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:287–301. [Google Scholar]
- 17.Hirschberg J, Cohen M, Harker M, Lotan T, Mann V, Pecker I. Pure Appl Chem. 1997;69:2151–2158. [Google Scholar]
- 18.Bouvier F, d'Harlingue A, Hugueney P, Marin E, Marion-Poll A, Camara B. J Biol Chem. 1996;271:28861–28867. doi: 10.1074/jbc.271.46.28861. [DOI] [PubMed] [Google Scholar]
- 19.Albrecht M, Klein A, Hugueney P, Sandmann G, Kuntz M. FEBS Lett. 1995;372:199–202. doi: 10.1016/0014-5793(95)00978-i. [DOI] [PubMed] [Google Scholar]
- 20.Badillo A, Steppuhn J, Deruère J, Camara B, Kuntz M. Plant Mol Biol. 1995;27:425–428. doi: 10.1007/BF00020196. [DOI] [PubMed] [Google Scholar]
- 21.Hugueney P, Römer S, Kuntz M, Camara B. Eur J Biochem. 1992;209:399–407. doi: 10.1111/j.1432-1033.1992.tb17302.x. [DOI] [PubMed] [Google Scholar]
- 22.Deruère J, Bouvier F, Steppuhn J, Klein A, Camara B, Kuntz M. Biochem Biophys Res Commun. 1994;199:1144–1150. doi: 10.1006/bbrc.1994.1350. [DOI] [PubMed] [Google Scholar]
- 23.Bouvier F, Hugueney P, d'Harlingue A, Kuntz M, Camara B. Plant J. 1994;6:45–54. doi: 10.1046/j.1365-313x.1994.6010045.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuntz M, Römer S, Suire C, Hugueney P, Weil J H, Schantz R, Camara B. Plant J. 1992;2:25–34. doi: 10.1111/j.1365-313x.1992.00025.x. [DOI] [PubMed] [Google Scholar]
- 25.Römer S, Hugueney P, Bouvier F, Camara B, Kuntz M. Biochem Biophys Res Commun. 1993;196:1414–1421. doi: 10.1006/bbrc.1993.2410. [DOI] [PubMed] [Google Scholar]
- 26.Hugueney P, Badillo A, Chen H C, Klein A, Hirschberg J, Camara B, Kuntz M. Plant J. 1995;8:417–424. doi: 10.1046/j.1365-313x.1995.08030417.x. [DOI] [PubMed] [Google Scholar]
- 27.Bouvier F, Keller Y, d'Harlingue A, Camara B. Biochim Biophys Acta. 1998;1391:320–328. doi: 10.1016/s0005-2760(98)00029-0. [DOI] [PubMed] [Google Scholar]
- 28.Mann V, Pecker I, Hirschberg J. Plant Mol Biol. 1994;24:429–434. doi: 10.1007/BF00024111. [DOI] [PubMed] [Google Scholar]
- 29.Pecker I, Gabbay R, Cunningham F X, Jr, Hirschberg J. Plant Mol Biol. 1996;30:807–819. doi: 10.1007/BF00019013. [DOI] [PubMed] [Google Scholar]
- 30.Ronen G, Cohen M, Zamir D, Hirschberg J. Plant J. 1999;17:341–351. doi: 10.1046/j.1365-313x.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- 31.Tanksley S D. In: Genetic Maps: Locus Maps of Complex Genomes. O'Brien S J, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 6.39–6.60. [Google Scholar]
- 32.Bernacchi D, Beck-Bunn T, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S D. Theor Appl Genet. 1998;97:381–397. doi: 10.1007/BF00223378. [DOI] [PubMed] [Google Scholar]
- 33.Fulton T M, Beck-Bunn T, Emmatty D, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S D. Theor Appl Genet. 1997;95:881–894. doi: 10.1007/BF00223378. [DOI] [PubMed] [Google Scholar]
- 34.Hurtado-Hernandez H, Smith P G. J Hered. 1985;76:211–213. [Google Scholar]
- 35.Lefebvre V, Kuntz M, Camara B, Palloix A. Plant Mol Biol. 1998;36:785–789. doi: 10.1023/a:1005966313415. [DOI] [PubMed] [Google Scholar]
- 36.Popovsky S, Paran I. Theor Appl Genet. 2000;101:86–89. [Google Scholar]
- 37.Fray R G, Grierson D. Plant Mol Biol. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- 38.Livingstone K D, Lackney V K, Blauth J R, Van Wijk R, Jahn M K. Genetics. 1999;152:1183–1202. doi: 10.1093/genetics/152.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn A. In: Methods in Molecular Biology. Cowell I G, Austin C A, editors. Vol. 69. Totowa, NJ: Humana; 1997. pp. 33–38. [Google Scholar]
- 40.Lessard P, Decroocq V, Thomas M. In: Plant Molecular Biology–A Laboratory Manual. Clark M S, editor. Berlin: Springer; 1997. pp. 179–181. [Google Scholar]
- 41.Michelmore R W, Paran I, Kesseli R V. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paran I, Aftergoot E, Shifriss C. Euphytica. 1998;99:167–173. [Google Scholar]
- 43.Popovsky S. Ph.D. thesis. Rehovot, Israel: Hebrew Univ. of Jerusalem; 1999. [Google Scholar]
- 44.Bird C R, Ray J A, Fletcher J D, Boniwell J M, Bird A S, Teulieres C, Blain I, Bramley P M, Schuch W. Bio/Technology. 1991;9:635–639. [Google Scholar]
- 45.Lancaster J E, Lister C S, Reay P F, Triggs C M. J Am Soc Hort Sci. 1997;122:594–598. [Google Scholar]
- 46.Davies B H, Matthews S, Kirk J T O. Phytochemistry. 1970;9:797–805. [Google Scholar]
- 47.Bonierbale M W, Plaisted R L, Tanksley S D. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bramley P, Teulieres C, Blain I, Bird C, Schuch W. Plant J. 1992;2:343–349. [Google Scholar]
- 49.Kinzer S M, Schwager S J, Mutschler M A. Theor Appl Genet. 1990;79:489–496. doi: 10.1007/BF00226158. [DOI] [PubMed] [Google Scholar]
- 50.Ben Chaim, A., Paran, I., Grube, R. C., Jahn, M. K., van Wijk, R. & Peleman, J. (2000) Theor. Appl. Genet., in press.
- 51.Giuliano G, Bartley G E, Scolnik P A. Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baroni Fornasiero R, Medeghini Bonatti P. J Plant Physiol. 1985;118:297–308. doi: 10.1016/S0176-1617(85)80189-9. [DOI] [PubMed] [Google Scholar]
- 53.Buckner B, Robertson D S. Methods Enzymol. 1993;214:311–323. doi: 10.1016/0076-6879(93)14074-s. [DOI] [PubMed] [Google Scholar]
- 54.Brown C R, Edwards C G, Yang C-P, Dean B B. J Am Soc Hort Sci. 1993;118:145–150. [Google Scholar]
- 55.Livingstone K D. Ph.D. thesis. Ithaca, NY: Cornell Univ.; 1999. [Google Scholar]
- 56.Zhang Y, Stommel J R. Theor Appl Genet. 2000;100:368–375. [Google Scholar]
- 57.Pillen K, Pineda O, Lewis C, Tanksley S. In: Genome Mapping in Plants. Paterson A H, editor. Austin, TX: Landes; 1996. pp. 281–307. [Google Scholar]
- 58.Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. Proc Natl Acad Sci USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]