Abstract
Overexpression of c-Myc in immortalized cells increases cell proliferation, inhibits cell differentiation, and promotes cell transformation. Recent evidence suggests that these effects, however, do not necessarily occur when c-Myc is overexpressed in primary mammalian cells. We sought to determine the immediate effects of transient overexpression of c-Myc in primary cells in vivo by using recombinant adenovirus to overexpress human MYC in mouse liver. Mice were intravenously injected with adenoviruses encoding MYC (Ad/Myc), E2F-1 (Ad/E2F-1), or β-galactosidase (Ad/LacZ). Transgene expression was detectable 4 days after injection. Expression of ectopic c-Myc was immediately accompanied by enlarged and dysmorphic hepatocytes in the absence of significant cell proliferation or apoptosis. These findings were not present in the livers of mice injected with Ad/E2F-1 or Ad/LacZ. Prominent hepatocyte nuclei and nucleoli were associated with the up-regulation of large- and small-subunit ribosomal and nucleolar genes, suggesting that c-Myc may induce their expression to increase cell mass. Our studies support a role for c-Myc in the in vivo control of vertebrate cell size and metabolism independent of cell proliferation.
The basic helix–loop–helix transcription factor, c-Myc, possesses the dual ability to transactivate genes that control cell proliferation and transrepress genes that induce growth arrest and differentiation (reviewed in refs. 1–3). c-Myc forms a heterodimeric complex with Max (4), and then binds to the enhancer box, 5′-CACGTG-3′ (5, 6), to activate transcription of genes. Overexpression of c-Myc in immortalized cells results in enhanced cell proliferation, inhibition of cell differentiation (7, 8), increased glucose metabolism (9), and anchorage-independent growth (10). In response to serum or glucose deprivation, c-Myc-overexpressing cells undergo apoptosis (11–14).
Identification of the genes regulated by c-Myc has helped clarify c-Myc function. Genuine c-Myc target genes must meet specific criteria to be classified as such (3), with a major criterion being that the protein encoded by the target gene is linked to cell proliferation or transformation. Genes found to be directly responsive to c-Myc include those involved in cell cycle regulation, such as cdc25A (15) and cyclins D1 and D2 (16), and those that control cell metabolism, such as ornithine decarboxylase (17) and lactate dehydrogenase A (9). However, the significance of target genes involved in protein translation, such as eIF-4E (18, 19) or MrDb, a putative RNA helicase (20), or carbamoyl-phosphate synthase transcarbamylase/aspartate dihydroorotase, an enzyme implicated in pyrimidine biosynthesis (21), have been less obvious, although it has been assumed that accelerated passage through G1 requires augmented RNA processing and protein synthesis (22).
It now appears that the increase in protein synthesis may serve another function beyond yet c-Myc-mediated cell proliferation. Recent studies implicate c-Myc in the control of cell growth (increase in cell mass), by a mechanism independent of its effect on the cell cycle. This impact on cell growth was most clearly demonstrated in Drosophila melanogaster, in which a dmyc mutation leads to diminutive flies that are slow to grow (23). In concordance with this observation, wing cells engineered to overexpress dmyc show an increase in cell mass, but not number (24). Similar observations have been made in mammalian cells. B cell-derived P493–6 cells that overexpress c-Myc have greater mass in every phase of the cell cycle (25). Transgenic mice that develop lymphoma as a result of the MYC transgene driven by the Eμ Ig enhancer, possess increased B cell size at all stages of B cell development regardless of cell-cycle phase (26). The identification of cell growth as a function of c-Myc overexpression has renewed interest in those genes involved in RNA processing and protein synthesis.
The finding that c-Myc induces cell growth in the absence of cell proliferation in some models points out a major deficiency in using immortalized or cancer cell lines to understand c-Myc function. Such systems may not accurately reflect its true function in mammalian cells in vivo, yet such information is vital for understanding its role in tumorigenesis. Contrary to the proliferative response immortalized cells display in response to c-Myc overexpression, primary mouse embryo fibroblasts that overexpress MYC undergo growth arrest from induction of the Mdm2 inhibitor, p19ARF (27). The differences between events in immortalized and primary cells undoubtedly reflect additional genetic alterations in the former that contribute to their immortality.
Studies involving c-Myc transgenic mice may be subject to similar criticism, because there is a high likelihood that compensatory genetic changes have occurred during development. Targeted expression of c-Myc in transgenic mice has resulted in lymphoma (28), liver tumor (29), or pancreatic tumor formation (30) as the mice age. Two recent studies have shed some light on the more immediate consequences of c-Myc function in vivo. One group examined the effects of deactivating a constitutively expressed MYC transgene in hematopoietic cells (31). Inactivation of the transgene led to regression of malignant T cell lymphomas and acute myeloid leukemias that arose from c-Myc overexpression. A second group developed a transgenic model that expressed an inducible c-Myc-estrogen receptor (MycER) chimera in the epidermis, which when exposed to 4-hydroxytamoxifen triggered the nuclear translocation of MycER, with development of keratinocyte proliferation and a precancerous epithelial lesion after 1–3 weeks of exposure (32). Deactivation of MycER caused regression of these lesions. Although this latter transgenic model most closely approximates c-Myc function in vivo, there was constitutive expression of the MycER chimera and nuclear localization of this protein even before exposure of the mice to 4-hydroxytamoxifen.
It is of profound interest to define the immediate consequences of c-Myc overexpression in the primary mammalian cell, because deregulated c-Myc expression is one of the earliest events in the multistep paradigm of carcinogenesis. We therefore sought to develop a model in which the immediate effects of transient overexpression of c-Myc in vivo could be assayed. The liver was chosen as the organ in which to study c-Myc function in vivo. c-Myc plays a critical role in the regenerative response of the liver after 70% partial hepatectomy, a stimulus for hepatocyte cell cycle entry (33, 34). Deregulated expression of c-Myc occurs in more than 50% of human hepatocellular carcinomas, suggesting that c-Myc participates in the pathogenesis of this tumor. Finally, transgene delivery to the liver, using recombinant adenovirus, has been well studied and at an appropriate viral titer is associated with minimal toxicity that could complicate interpretation of c-Myc's effects. Because hepatocytes could be transduced with high efficiency and transgene expression occurs within 3–5 days of delivery, the advantage of this system was that we could study the effects of c-Myc expression in the normal cell in the absence of major compensatory genetic changes. By using recombinant adenovirus to deliver the human MYC transgene to murine liver, we found that ectopic expression of c-Myc resulted in a novel phenotype of hepatocyte hypertrophy with concomitant enlargement of nuclei and nucleoli. These effects were specific to c-Myc and not observed with the overexpression of E2F-1, a transcription factor that regulates cell proliferation. Cell hypertrophy as a result of c-Myc overexpression was accompanied by an increase in the expression of genes encoding ribosomal and nucleolar proteins, but occurred in the absence of significant proliferation. Our data underscore the fact that the effect of c-Myc in primary cells cannot be predicted from prior observations made in immortalized cells.
Materials and Methods
Adenoviral in Vivo Gene Transfer.
Animal studies were approved by The Johns Hopkins University School of Medicine Animal Use and Care Committee. Adenoviral constructs Ad/LacZ and Ad/Myc were the generous gifts of W. El-Deiry (University of Pennsylvania, Philadelphia) and have been described (35). Ad/E2F-1 was the gift of J. R. Nevins (Duke University, Durham, NC) (36). Two-month-old male BALB/c mice, weighing 20–25 g, were intravenously injected in a volume of 200 μl with 4 × 109 plaque-forming units (pfu) of Ad/LacZ, Ad/Myc, or Ad/E2F-1. Mice were killed by CO2 on days 3, 4, and 5 after adenoviral injection. Livers were quickly isolated and frozen for subsequent experiments.
β-Galactosidase Assay.
Day 3, 4, and 5 livers transduced with Ad/LacZ were frozen in Tissue-Tek OCT compound (Sakura) and sectioned onto glass slides. The frozen sections were fixed by incubating the slides in PBS containing 2% formaldehyde for 5 min at 4°C. The sections were washed twice with PBS and assayed for β-galactosidase activity by incubating the slides in PBS containing 5 mM potassium ferricyanate, 2 mM MgCl2 and (1/20) vol of 20 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside in dimethylformamide at 4°C. After 6 h, positive staining was visualized, and the reaction was stopped by washing the slides with PBS.
Western Analysis.
Ten milligrams of frozen liver tissue was minced by using a razor blade at 4°C. The tissue was transferred into a 0.5-ml glass tube containing hot 2× Laemmli buffer (95°C) (0.5 M Tris⋅Cl, pH 6.8/20% glycerol/4% SDS/10% β-mercaptoethanol/2.5% bromphenol blue) and homogenized by using a glass pestle. The lysate was then incubated at 95°C for 5 min and spun at 14,000 rpm at 25°C for 5 min to spin down the cellular debris. The supernatant was loaded into 12% SDS/PAGE gel and transferred to polyvinylidene difluoride membrane (Millipore). The c-Myc protein was detected by using 9E10 antibody as described (37).
Northern Analysis.
Total RNA was prepared by using TRIzol reagent (Life Technologies), and 20 μg of total RNA was loaded per lane on a 1.2% agarose/formaldehyde gel and transferred to nylon membrane (Schleicher & Schuell). The cDNAs used for probes were the generous gifts of Q. B. Guo (National Institutes of Health, Bethesda, MD), and 32P-labeled probes were generated by using the PrimeIt-II random primer labeling kit (Stratagene). The hybridization and washing steps were carried out as described (37), and the positive signals were visualized by using a PhosphorImager and autoradiography.
Apoptosis Assay.
Apoptosis was detected by using terminal deoxyribonucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining. Tissue sections were deparaffinized by three 5-min washes in xylene, two 5-min washes in 100% ethanol, one 3-min wash in 95% ethanol, and one 3-min wash in 70% ethanol, followed by one 5-min wash in PBS. TUNEL staining was performed by using the ApopTag kit (Intergen). Positive-staining cells were visualized by using a fluorescence microscope with the excitation filter set at 488 nm (Nikon).
Immunohistochemistry.
Ki-67 staining was performed as described (38), with the modification that the antibody incubation was done at room temperature for 30 min by using a 1:200 dilution of Ki-67 antibody (MIB5, Zymed). For methyl green pyronin staining, the paraffinized sections were rinsed with deionized H2O and then acetate buffer (pH 4.8). Sections were stained by incubation in a buffer containing 2% aqueous methyl green, 2% aqueous pyronin Y (or G), and 28% glycerol (pH 4.8) for 30 min at room temperature. The sections were blot-dried and rinsed with equal parts of acetone/xylene and then xylene.
Cell Morphometrics.
Image analysis of hematoxylin/eosin-stained liver sections was performed by using imagepro software. Cells were viewed under ×40 magnification, and every hepatocyte in each field containing a nucleus was used to calculate the average number of hepatocytes per high power field and average area of hepatocytes. Cell volumes were derived from the area measurements.
Results
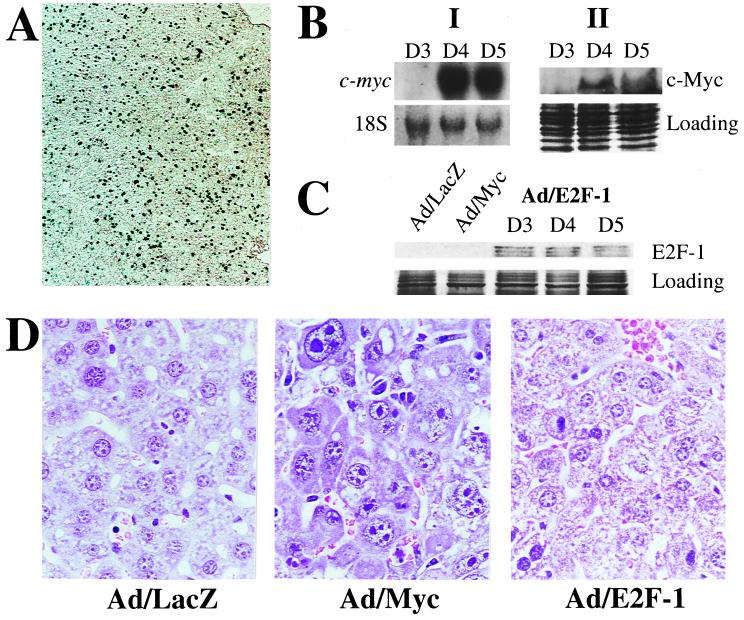
To study the effect of exogenous overexpression of c-Myc in the liver, we injected BALB/c mice intravenously with 4 × 109 pfu of Ad/Myc. Control mice were injected with Ad/LacZ. Mice killed on days 4 and 5 had livers that stained positively for β-galactosidase activity, with 40% of hepatocytes staining positive by day 5 after injection (Fig. 1A). MYC transgene and protein expression were detectable by day 4 postinjection (Fig. 1B).
Figure 1.
(A) β-Galactosidase staining of day 5 Ad/LacZ liver. (B) (I) Northern blot showing exogenous MYC gene expression starting on day 4. 18S is shown as loading control. (II) Western blot showing c-Myc protein expression in Ad/Myc livers. Coomassie staining is shown for loading control. D3, D4, and D5 indicate total RNA and protein samples isolated from Ad/Myc livers killed on days 3, 4, and 5, respectively. (C) Western blot showing that E2F-1 is expressed by day 3 postinjection of recombinant adenovirus carrying E2F-1. E2F-1 is not detectable in Ad/LacZ or Ad/Myc livers. (D) Hematoxylin/eosin staining of day 4 Ad/LacZ, Ad/Myc, and Ad/E2F-1 liver. (Magnification: A, ×5; D, ×40.)
Normal hepatic architecture was disrupted by the presence of enlarged, dysmorphic hepatocytes in livers from Ad/Myc mice killed on day 4 (Fig. 1D); such changes were absent in Ad/LacZ livers. No significant inflammatory infiltrate in any liver section was observed. To determine whether cell enlargement could be induced by another transcription factor known to regulate cell proliferation, we expressed E2F-1 in mouse liver after injecting the same adenovirus titer used for Ad/LacZ and Ad/Myc. E2F-1 expression was seen in mouse livers 3 days after injection (Fig. 1C), yet no increase in hepatocyte size was observed (Fig. 1D). Hence, the cell enlargement in Ad/Myc livers appears to be c-Myc specific because E2F-1 failed to increase cell size despite its recognized role in cell proliferation.
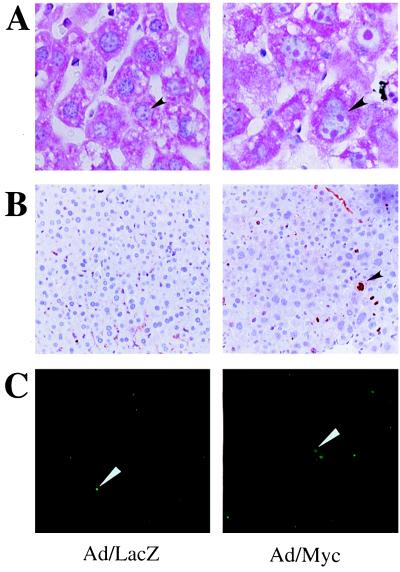
Morphometrics of 80 hepatocytes from several random high- power fields of both Ad/Myc and Ad/LacZ livers determined that the average volume of Ad/Myc hepatocytes [819 ± 68 arbitrary units (average ± SE), n = 82 cells] was 2.0 times greater than that from Ad/LacZ mice (399 ± 14 arbitrary units, n = 78 cells). In other terms, the average number of hepatocytes per high-power field was 27 ± 1 for Ad/Myc livers compared with 39 ± 4 for Ad/LacZ livers. In addition, both nuclei and nucleoli were enlarged in Ad/Myc livers. Nucleoli were readily distinguished from condensed chromatin because they stained red by methyl green pyronin, which stains RNA red and DNA blue (Fig. 2A).
Figure 2.
(Left) Day 4 Ad/LacZ liver. (Right) Day 4 Ad/Myc liver. (A) Methyl green pyronin staining of the sections. The arrows highlight enlarged nucleoli of Ad/Myc hepatocytes. (B) Ki-67 staining of liver sections. The arrow indicates positive staining of a proliferating cell. (C) TUNEL staining of liver sections. The arrows indicate nuclei of cells undergoing apoptosis. (Magnification: A, ×60; B and C, ×10.)
We next sought to determine whether the enlarged hepatocytes were proliferating or undergoing apoptosis. Liver sections were stained with an antibody to the cell proliferation marker, Ki-67. Although there was 2.5-fold greater staining for Ki-67 in Ad/Myc liver compared with Ad/LacZ liver, fewer than 0.03% hepatocytes in Ad/Myc liver stained positively for Ki-67 (Fig. 2B), indicating that the vast majority of enlarged hepatocytes were not proliferating. A TUNEL apoptotic assay of liver sections showed that despite an increase in the number of apoptotic cells in Ad/Myc liver (7%) compared with that in Ad/LacZ liver (2%), the majority of enlarged hepatocytes in Ad/Myc liver were not undergoing apoptosis (Fig. 2C). Taking these results together, we concluded that the hepatocyte enlargement represents cell hypertrophy.
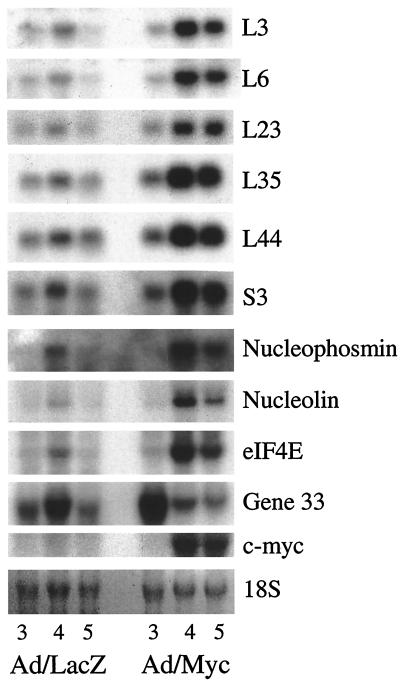
The prominent nucleoli led us to consider whether expression of ribosomal genes was altered, because nucleolar size is indicative of ribosomal RNA synthesis (39). Northern analysis indicated that several ribosomal genes and genes that encode proteins engaged in ribosome biogenesis and protein translation are indeed up-regulated in Ad/Myc liver compared with Ad/LacZ liver (Fig. 3). Nucleolin, recently reported to be transcriptionally regulated by c-Myc (40), displayed enhanced expression on day 4. The up-regulation of ribosomal gene expression does not reflect a universal effect on all gene expression, because gene 33, a hormonally regulated gene activated in cell proliferation and liver regeneration, is down-regulated in Ad/Myc livers. Gene 33 was previously identified as a gene down-regulated in the livers of transgenic mice engineered to overexpress c-Myc in the liver (unpublished data). We recently used poly(A)-RNA isolated from both Ad/Myc and Ad/LacZ liver to generate cDNA probes that were hybridized to arrays containing more than 8,700 cDNAs so that we could identify additional differentially expressed genes. Only 227 genes (2.5%) were found to be greater than 1.8-fold differentially expressed (data not shown), indicating that the expression levels of only a small number of genes change in the liver in response to c-Myc. Thus, the genes whose expression we have identified as altered in this model are likely to be significant.
Figure 3.
Northern blot showing the expression levels of ribosomal and nucleolar genes in Ad/LacZ and Ad/Myc livers. All of the ribosomal genes examined show increased expression after the ectopic MYC expression. Expression of the immediate-early gene 33 is down-regulated in Ad/Myc liver. 18S is shown as loading control. Numbers at the bottom indicate the days mice were killed after adenovirus injection.
Discussion
To better understand the function of c-Myc in vivo, we developed a murine model in which we used recombinant adenovirus to deliver the human MYC transgene to the liver. There was avid hepatic uptake of the recombinant adenovirus and rapid expression of the transgene by the fourth day after delivery. Expression of the transgene was accompanied by marked enlargement of hepatocytes, most of which were not proliferating or undergoing apoptosis. The increase in cell size could not be attributed to the adenoviral vector itself, as Ad/LacZ failed to induce hypertrophy. Because MYC products have been reported to transactivate an adenoviral promoter (41), we also considered that c-Myc could complement the adenoviral E1A defect in the recombinant Ad/Myc, allowing expression of other adenoviral proteins. However, expression of E2A, which is induced by E1A, was not detected in Ad/Myc liver (data not shown), and therefore cell hypertrophy could not be attributed to expression of adenoviral proteins.
c-Myc-dependent cell hypertrophy occurs in the absence of cell proliferation in the liver, as has been reported in a few other models (23, 25, 26). A similar effect was not observed with the E2F-1 transgene delivery to the liver, suggesting that the effect is specific to c-Myc and not generalized to all transcription factors that affect cell proliferation. The absence of increased hepatocyte mass in response to E2F-1, which activates genes that control S phase progression, is consistent with prior observations that E2F-1 failed to increase cell mass when overexpressed in Drosophila wing cells (24). Thus, cell proliferation and cell hypertrophy are events that can indeed be segregated.
c-Myc appears to induce cell and nucleolar enlargement by altering the expression of genes that enhance protein synthesis. We found that the expression of several ribosomal subunit proteins, translation initiation factors, and nucleolar proteins increased in Ad/Myc liver in concordance with c-Myc overexpression. The up-regulation of such genes may explain why we observed very prominent nucleoli in addition to enlarged cells. These findings are consistent with prior observations that genes encoding proteins involved in protein translation, such as the RNA helicase, MrDb, and the translation initiation factors, eIF-4E and eIF-2A, are up-regulated by c-Myc in several systems (18–20). Furthermore, nucleolin, responsible for ribosomal biogenesis, and BN51, a cofactor of RNA polymerase III, were recently identified as c-Myc-responsive genes (40).
The relationship between c-Myc and protein synthesis is intriguing and reflects another effect of c-Myc on cell metabolism. c-Myc is likely to increase cell mass simply by its effect on nucleolar and ribosomal protein expression. The nucleolus possesses several functions, chief of which is ribosomal synthesis (42). Studies in D. melanogaster indicate that nucleolar proteins involved in ribosome assembly appear critical for maintaining cell and organismal size. Thirteen of the Drosophila minute loci associated with small body size and short bristles arise from mutations to ribosomal genes (43). Partial loss of function of the gene mfl, which encodes a ubiquitous nucleolar protein that participates in ribosomal RNA processing and pseudouridylation, results in minifly, yet another small Drosophila mutant (44).
The absence of significant proliferation in response to c-Myc overexpression in vivo contrasts with the effect in immortalized cell lines, which proliferate in response to ectopic c-Myc over expression. Studies of hyperplasia versus hypertrophy in other cells, such as vascular endothelium, lead us to surmise that hypertrophy in our model results from overexpressing c-Myc in growth-arrested hepatocytes as opposed to ones undergoing active proliferation (45, 46). Ectopic expression of c-Myc appears unable to overcome the signals that keep normal hepatocytes in G0, and, therefore, cells do not proliferate in response to acute overexpression of c-Myc.
Cell hypertrophy and induction of nucleolar proteins may relate to cell transformation, because tumor cells characteristically possess enlarged nucleoli. Nucleolin staining is typically increased (47) and has been used as a tumor marker. It remains to be determined whether the gene expression patterns induced by c-Myc in increasing cell growth are distinct from those implicated in cell proliferation. As Elend and Eilers (48) eloquently discussed, Myc induces cyclin D2 even in the presence of cycloheximide (16), indicating that protein synthesis, obviously required for cell growth, is not required for cell proliferation. Cell proliferation and apoptosis are closely linked in immortalized cells that overexpress c-Myc, but in our model, enhanced cell size did not trigger overwhelming apoptosis. Thus, the genes that are c-Myc-responsive and govern c-Myc phenotypes in mammalian cells in vivo may differ substantially from those identified in vitro.
Acknowledgments
We thank L. Womack for technical assistance, J. Boitnott for histological interpretation, I. Simon for assistance with morphometric analysis, and W. Gage for Ki-67 staining. We also thank Dr. Gary Ketner for providing the E2A probe. This work was funded by National Institutes of Health Grants CA64258 (to L.A.L.) and CA57341 (to C.V.D.).
Abbreviations
- Ad/Myc
adenovirus encoding human MYC
- Ad/E2F-1
adenovirus encoding E2F-1
- Ad/LazZ
adenovirus encoding β-galactosidase
- MycER
Myc-estrogen receptor
- pfu
plaque-forming units
- TUNEL
deoxyribonucleotidyl transferase-mediated dUTP nick end labeling
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200372597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200372597
References
- 1.Cole M D, McMahon S B. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 2.Dang C V, Resar L M, Emison E, Kim S, Li Q, Prescott J E, Wonsey D, Zeller K. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 3.Grandori C, Eisenman R. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood E M, Lüscher B, Eisenman R N. Genes Dev. 1992;6:71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- 5.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F W, Eisenman R N, Weintraub H. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freytag S O. Mol Cell Biol. 1988;8:1614–1624. doi: 10.1128/mcb.8.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachman H M, Skoultchi A L. Nature (London) 1984;318:592–594. doi: 10.1038/310592a0. [DOI] [PubMed] [Google Scholar]
- 9.Shim H, Dolde C, Lewis B C, Wu C, Dang G, Jungmann R A, Dalla-Favera R, Dang C V. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone J, de Lange T, Ramsay G, Jokobovits E, Bishop J M, Varmus H E, Lee W M F. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amati B, Littlewood T, Evan G, Land H. EMBO J. 1993;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Askew D, Ashmun R, Simmons B, Cleveland J. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 13.Evan G, Wyllie A, Gilbert C, Littlewood T, Land H, Brooks M, Waters C, Penn L, Hancock D. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 14.Shim H, Chun Y S, Lewis B C, Dang C V. Proc Natl Acad Sci USA. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galaktionov K, Chen X, Beach D. Nature (London) 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 16.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bello-Fernandez C, Packham G, Cleveland J L. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenwald I, Rhoads D, Callanan L, Isselbacher K, Schmidt E. Proc Natl Acad Sci USA. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones R, Branda J, Johnston K, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt E. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandori C, Mac J, Siebelt F, Ayer D, Eisenman R. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 21.Miltenberger R J, Sukow K A, Farnham P J. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt E V. Oncogene. 1999;18:2988–2996. doi: 10.1038/sj.onc.1202751. [DOI] [PubMed] [Google Scholar]
- 23.Johnston L, Prober D, Edgar B, Eisenman R, Gallant P. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neufeld T P, de la Cruz A F A, Johnston L A, Edgar B A. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 25.Schuhmacher M, Staege M, Pajic A, Polack A, Weidle U, Bornkamm G, Eick D, Kohlhuber F. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 26.Iritani B, Eisenman R. Proc Natl Acad Sci USA. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langdon W, Harris A, Cory S, Adams J. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 29.Sandgren E P. Oncogene. 1989;4:715–724. [PubMed] [Google Scholar]
- 30.Sandgren E, Quaife C, Paulovich A, Palmiter R, Brinster R. Proc Natl Acad Sci USA. 1991;88:93–97. doi: 10.1073/pnas.88.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsher D W, Bishop J M. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 32.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Mol Cell. 1999;3:565–567. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 33.Fausto N, Shank P. Hepatology. 1983;3:1016–1023. doi: 10.1002/hep.1840030621. [DOI] [PubMed] [Google Scholar]
- 34.Haber A H, Mohn K, Diamond R, Taub R. J Clin Invest. 1993;91:1319–1326. doi: 10.1172/JCI116332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell K O, El-Deiry W S. Cell Growth Differ. 1999;10:223–230. [PubMed] [Google Scholar]
- 36.DeGregori J, Leone G, Ohtani K, Miron A, Nevins J R. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 37.Lewis B, Shim H, Li Q, Wu C, Lee L, Maity A, Dang C. Mol Cell Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King G, Payne S, Walker F, Murray G I. J Pathol. 1997;183:237–241. doi: 10.1002/(SICI)1096-9896(199710)183:2<237::AID-PATH893>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Frank D, Roth M. J Cell Biol. 1998;140:1321–1329. doi: 10.1083/jcb.140.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greasley P J, Amati B C. Nucleic Acids Res. 2000;28:446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onclercq R, Gilardi P, Lavenu A, Cremisi C. J Virol. 1988;62:4533–4537. doi: 10.1128/jvi.62.12.4533-4537.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambertsson A. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- 44.Giordano E, Peluso I, Senger S, Furia M. J Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neufeld T, Edgar B. Curr Opin Cell Biol. 1998;10:784–790. doi: 10.1016/s0955-0674(98)80122-1. [DOI] [PubMed] [Google Scholar]
- 46.Braun-Dullaeus R C, Mann M J, Ziegler A, von der Leyen H E, Dzau V J. J Clin Invest. 1999;104:815–823. doi: 10.1172/JCI5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs R. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 48.Elend M, Eilers M. Curr Biol. 1999;9:R936–R938. doi: 10.1016/s0960-9822(00)80109-8. [DOI] [PubMed] [Google Scholar]