Abstract
The solution structure of bacteriocin AS-48, a 70-residue cyclic polypeptide from Enterococcus faecalis, consists of a globular arrangement of five α-helices enclosing a compact hydrophobic core. The head-to-tail union lies in the middle of helix 5, a fact that is shown to have a pronounced effect on the stability of the three-dimensional structure. Positive charges in the side chains of residues in helix 4 and in the turn linking helix 4 to helix 5 form a cluster that most probably determine its antibacterial activity by promoting pore formation in cell membranes. A similar five-helix structural motif has been found in the antimicrobial NK-lysin, an effector polypeptide of T and natural killer (NK) cells. Bacteriocin AS-48 lacks the three disulfide bridges characteristic of the saposin fold present in NK-lysin, and has no sequence homology with it. Nevertheless, the similar molecular architecture and high positive charge strongly suggest a common mechanism of antibacterial action.
Keywords: cationic antibacterial peptides, NMR solution structure, five-helix globule, cyclic polypeptide, membrane permeation
Considerable effort has been invested in recent years toward the use of cationic antibacterial peptides as an alternative to conventional antibiotics. Interest in these peptides has been fuelled because of the emergence and widespread diffusion of drug-resistant bacteria, fungi, and parasites, a problem of great concern especially in clinical settings. These peptides usually exert a potent antimicrobial activity by mechanisms other than enzyme inhibition, so that one of the forms of developing resistance, that of altering the drug's target, does not apply, and the emergence of resistant strains is made more difficult.
We have been working in recent years on one of these peptides, the bacteriocin AS-48, a 70-residue cyclic peptide produced by Enterococcus faecalis S-48, which shows a broad antimicrobial spectrum against both Gram-positive and Gram-negative bacteria (1–3). Bacteriocin AS-48 is encoded by the 68-kb pheromone-responsive plasmid pMB2 (4), and the gene cluster involved in production and immunity has been identified (5). This peptide exerts a bactericidal action on sensitive cells (2), and its target is the cytoplasmic membrane, in which it opens pores, leading to the dissipation of the protonmotive force and cell death (6), a mechanism similar to that proposed for the action of defensins or, most generally, cationic antibacterial peptides (7).
The amino acid composition of purified AS-48 (8) shows the absence of lanthionine, β-methyllanthionine, and dehydrated residues, making it clearly different from lantibiotics. Bacteriocin AS-48 also differs from defensins in that it does not contain cysteines and consequently its structure is not stabilized by disulfide bridges. Composition analysis shows a high proportion of basic to acidic amino acids, conferring to this peptide a strong basic character, with a pI close to 10.5.
Elucidation of the primary structure of AS-48 was hindered initially by its resistance to Edman degradation, indicating that the N terminus was blocked. Also, treatment with carboxypeptidases A and B did not cleave off any C-terminal residues, suggesting that it may be a cyclic peptide. Further determination of the AS-48 structural gene DNA sequence (9), together with the sequences of AS-48 protease digestion fragments and mass spectrometry determinations (10), allowed us to determine unambiguously the cyclic structure of the molecule. To our knowledge, this represents the first example of a posttranslational modification in which a cyclic structure arises from a “tail-to-head” linkage of the gene product. In addition to the lack of stabilizing disulfide bridges, the cyclic character of this polypeptide is a further unusual feature that distinguishes AS-48 from the rest of cationic antibacterial peptides described until now.
In a previous work, we have reported the complete NMR assignment of the proton resonances of AS-48 together with the resulting secondary structure pattern (11). The detailed mechanism of its interaction with cell membranes and, as a final objective, the understanding of its killing of target cells require, as a fundamental step, the knowledge of its three-dimensional structure, a subject that has been the main objective of the present work. The resulting three-dimensional structure of bacteriocin AS-48 consists of a globular arrangement of five α-helices enclosing a compact hydrophobic core. The global fold is highly similar to the one found for NK-lysin, an antibacterial and tumorolytic 78-residue polypeptide of T and natural killer (NK) cells, isolated from porcine small intestine (12). Despite the lack of sequence homology, the structural similarity between the two polypeptides strongly suggests a common mechanism of antibacterial action. A different orientation of the direction of maximum electropositive potential with respect to the five-helix bundle is observed, which could mean a different initial approach and topological binding of the polypeptides to the target cell membranes.
Methods
Sedimentation Equilibrium.
Sedimentation equilibrium experiments were performed in a Beckman Optima XL-A ultracentrifuge using a Ti60 rotor and double sector centerpieces of Epon–charcoal (optical pathlength 12 or 4 mm, depending on peptide concentration). Samples of AS-48 in the concentration range 1.5–0.35 mM and equilibrated in water, with or without 100 mM NaCl, were centrifuged at 30,000 rpm and 20°C. Radial scans at various wavelengths (280–300 nm) were taken every 2 h until there were no differences between two successive scans (equilibrium conditions). The weight-average molecular mass (Mw) of AS-48 was determined by using the programs xlaeq and eqassoc (Beckman) with the partial specific volume of AS-48 set to 0.732 ml/g at 20°C as calculated from its amino acid composition (13).
Electrostatic repulsion between like-charged molecules at low ionic strength and high peptide concentrations would lead to nonideal behavior. Under these conditions the radial distribution of solute sedimentation equilibrium leads to the following expression for the apparent weight-average molecular mass, Mw,app (13, 14):
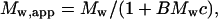 |
1 |
where B is the term for nonideality and c is the concentration.
For a monomer–dimer equilibrium, Mw is related to the monomer molecular mass, M1, as follows (14, 15):
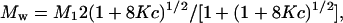 |
2 |
where K is the equilibrium constant in molar units, and c is the total concentration of AS-48.
Values of equilibrium constant and B for the low ionic strength experiments were calculated by fitting the data to Eqs. 1 and 2, using a nonlinear least-squares procedure, based in a modified Nelder–Mead simplex algorithm (16).
NMR Spectroscopy.
Bacteriocin AS-48 was purified and NMR samples were prepared as described previously (11). NMR experiments were recorded with a 1.5 mM sample of AS-48 containing 0.1 M NaCl in the presence of either 2H2O or 90% H2O/10% 2H2O. All spectra were acquired at 298 K and pH 3.0 (not corrected for isotope effects) in the phase-sensitive mode with time-proportional phase incrementation mode (17) on a Bruker AMX-600 spectrometer. 1H homonuclear correlated spectroscopy (COSY) (18), total correlation spectroscopy (TOCSY) (19), and nuclear Overhauser enhancement (NOE) spectroscopy (NOESY) (20) spectra with mixing times of 80 and 150 ms were recorded by standard methods with water suppression achieved by selective presaturation of the water signal or by including the WATERGATE module (21) in the original pulse sequences. 1H NMR resonances were assigned (11) by using standard sequential assignment procedures (22).
Collection of Structural Constraints.
Nonsequential and long-range NOE cross-peaks were assigned in consecutive rounds of peak assignments and distance geometry calculations. Possible assignments for NOE cross-peaks were obtained by using the program xeasy (23), and their intensities were determined by using the integration routines of the same program. The intensities were converted into upper distance bounds with the program caliba (24). Cross-peaks not integrated because of partial overlap were qualitatively classified as strong, medium, and weak and assigned distance constraints of 3.0, 4.0, and 5.0 Å, respectively. The program dyana 1.5 (25) was used to obtain stereospecific assignments from structures obtained in the final stages of the distance geometry calculations. For protons not stereospecifically assigned, and methyl and aromatic protons, the usual pseudoatom corrections were applied (22). No explicit hydrogen bonds were used in the structure calculations.
Structure Calculation.
Initial structures for bacteriocin AS-48 were calculated from the NMR-derived upper bounds by using the variable target function method implemented in the program dyana 1.5 starting from random structures. Because of the cyclic nature of the protein, constraints corresponding to a normal peptide bond had to be added to the new head-to-tail covalent bond.
The best 20 structures obtained in the final round of dyana calculations were further refined by restrained molecular dynamics and energy minimization calculations with the gromos package (26). The structures were first energy-minimized and then submitted to 50 ps of molecular dynamics at 300 K. During the dynamics, the integration step was 2 fs, and a time constant for coupling to the bath of 0.1 ps and a distance restraint constant of 70 kJ⋅mol−1⋅Å−2 were used. The last 5 ps of the trajectory were used for averaging, and the resulting structures were finally energy minimized. The input constraints for the structure calculation have been also deposited in the Brookhaven Protein Data Bank. The programs MolMol (27), molscript (28), and vadar (29) were used in structure analysis, molecular graphics manipulations, and surface area calculations.
Results
Sedimentation Equilibrium.
The association state of the sample of AS-48 under the conditions used in the NMR experiments was tested by equilibrium ultracentrifugation, as detailed in Methods. At low-salt conditions, the Mw of AS-48 decreases with the concentration, which is a sign of nonideality (see Methods). The best fit parameter values of the performed analysis were the following: M1 = 7,200 g⋅mol−1 (kept constrained in the fitting); K = 4.0 × 10−56 M−1; and B = 0.31 (nonideal term). The very low value of the association constant definitely indicates that AS-48 is entirely monomeric under the experimental conditions studied. However, the large value of the B term means that, in those conditions, electrostatic repulsion between like-charged molecules lead to a very significant nonideal behavior. Data at 100 mM NaCl showed the absence of nonideality and provided a value for Mw close to the theoretical monomer molecular mass (M1 = 7,200 g⋅mol−1).
Structure Calculations of AS-48.
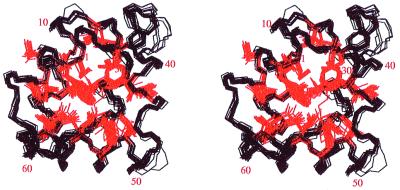
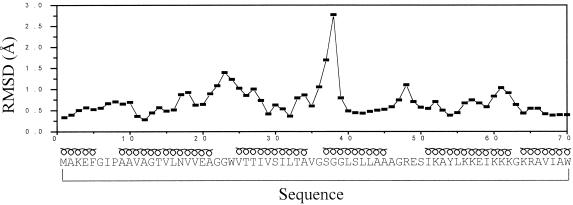
Solution structures were determined by using the procedures described in Methods. The number of structurally relevant intraresidual, sequential, and medium- and long-range constraints are summarized in Table 1. The total number used for the final structure calculations was 784. The restraints are well distributed all over the structure, as expected for a globular protein. The NMR structure of bacteriocin AS-48 protein is represented by 20 conformers with an average pairwise RMSD of 0.8 Å and 1.4 Å for the backbone and all heavy atoms, respectively (Table 1). The backbone conformations of the 20 structures of the cyclic polypeptide are shown superimposed in Fig. 1. As usual, the regions corresponding to elements of regular secondary structure, α-helical conformation in this case, are best defined. The loops connecting the different helices are also well defined, with the exception of the segment Gly-Ser-Gly-Gly, residues 36–39, which presents two different families of conformations, and whose atoms exhibit the largest local RMSD (see Fig. 2). When this region is excluded from the comparison, the average pairwise backbone RMSD drops to 0.7 Å. All of the backbone torsion angles for the non-glycine residues fall in the allowed region of the φ, ψ Ramachandran plot for all of the structures with the exception of Ser-50, which persistently shows a positive φ angle, and Ser-37 in 4 of 20 structures. Atomic coordinates for the 20 converged structures have been deposited in the Brookhaven Protein Data Bank with accession code 1e68.
Table 1.
Summary of NMR structure calculations
| Type of constraints | Number of distance
constraints
|
|||
|---|---|---|---|---|
| Total | <3.5 Å | 3.5–4.5 Å | ≥4.5 Å | |
| Intraresidual (|i − j| = 0) | 271 | 115 | 110 | 46 |
| Sequential (|i − j| = 1) | 249 | 66 | 107 | 76 |
| Medium-range (2 ≤ |i − j| ≤ 5) | 131 | 8 | 73 | 50 |
| Long-range (|i − j| > 5) | 133 | 1 | 36 | 96 |
| All | 784 | 190 | 326 | 268 |
| Residual constraint violations | Sum* | Maximum | ||
| Upper limits, Å | 15.5 | 0.6 | ||
| Energy terms, kJ·mol−1 | Avg* | Range | ||
| Total energy | −1,278 | −1,640 to −1,011 | ||
| Lennard–Jones energy | −2,266 | −2,364 to −2,099 | ||
| NOE term | 109 | 93 to 146 | ||
| Pairwise RMSD, Å | Backbone* | All heavy atoms* | ||
| All residues (1–70) | 0.8 ± 0.3 | 1.4 ± 0.3 | ||
| Excluding residues 36–39 | 0.6 ± 0.2 | 1.3 ± 0.2 | ||
| Buried residues | 0.5 ± 0.2 | 0.7 ± 0.2 | ||
| Surface residues | 0.9 ± 0.3 | 1.6 ± 0.3 | ||
RMSD, root-mean-square deviation.
Average values over the 20 computed structures.
Figure 1.
Stereoscopic view of the superposition of the 20 best structures of bacteriocin AS-48, resulting from the DYANA calculation. In black: superposition of backbone atoms. In red: superposition of hydrophobic side chains at the core of the structure. The figure was drawn with the program MOLSCRIPT.
Figure 2.
Pairwise RMSD of the coordinates of the backbone atoms of the 20 best calculated structures vs. sequence. α-Helical regions are indicated above the sequence with a loopy symbol. The Gly-rich segment, Gly-Ser-Gly-Gly, residues 36–39, presents two different families of conformations, which is the origin of the high RMSD value in that region.
Description of the Structures.
Bacteriocin AS-48 folds in an all α-helix manner with the five α-helices arranged according to the topology indicated in Figs. 3 and 4. The five helices, named α1 to α5, span residues 9–21, 25–34, 37–45, 51–62, and 64–5 (the amino acid sequence and an illustration of the spanning of the helices is given in the horizontal axis of Fig. 2). Helices α1 and α2 run antiparallel to each other. α4 is almost perpendicular to α1 and α2, and it runs parallel to the plane defined by them. α3 is perpendicular to α4 and intersects the plane defined by α1 and α2 at an angle of around 40°. α5 closes the helix bundle by running almost perpendicular to α3 and forming an angle of 50° with α4 and α1. The global structure is very compact, with the hydrophobic sides of the α-helices forming the core of the protein (see Fig. 1).
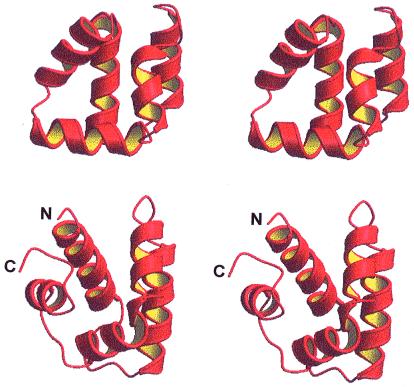
Figure 3.
A comparison of the three-dimensional structures of bacteriocin AS-48 (Upper) and NK-lysin (Lower). The largest difference concerns helix 5. A virtual bond between the N and C termini to make NK-lysin a cyclic polypeptide would increase the similarity of the structures of the two peptides.
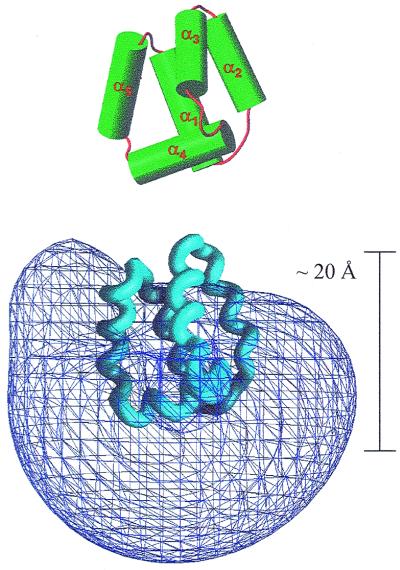
Figure 4.
(Upper) AS-48, with α-helices represented as cylinders. (Lower) Positive electrostatic potential at the level of 2kT/q, as calculated with the program GRASP. The direction of maximum change of the potential is approximately vertical. The length of the polypeptide in that orientation, 20 Å, is given so as to be compared with 40 Å, the standard thickness of a lipid bilayer.
The backbone hydrogen bonding pattern is essentially the one suggested on the basis of the previously determined secondary structure (11). Thus, most of the expected hydrogen bonds for the helical segments have been found. The five α-helices have, in general, a regular internal hydrogen bonding pattern, characterized by repeated hydrogen bonds between the CO of residue i and the NH of residue i + 4. In addition, some hydrogen bonds are involved in the stabilization of the loops connecting the α-helices. Thus, the hydrogen bonds between Ala-2 and Ile-7, Val-18 and Gly-23, and Thr-33 and Gly-36 stabilize the loops between helices α5 and α1, α1 and α2, and α2 and α3, respectively. No hydrogen bonds between residues located very far in the sequence are detected, with the exception of the hydrogen bond between the backbone carbonyl oxygen of Phe-5 and the side-chain hydroxylic proton of Ser-50. Twenty-nine residues, mostly those belonging to the internal hydrophobic core, showed a well-defined configuration (RMSD < 1 Å) for the entire side chain.
Search for Similar Folds.
The program dali (30) was used to search for structurally similar proteins in the Protein Data Bank. As expected for an all-helical-domain polypeptide, many DNA-binding proteins, containing helix–turn–helix motifs, were detected that showed some structural similarity with AS-48. However, these similarities did not extend to all five α-helices of AS-48, an observation that, together with the absence of biological data supporting any implication of AS-48 in DNA binding, makes the found scores irrelevant. More interesting are the structural similarities found with other proteins that interact with cell membranes and contain helical repeats, such as the potassium channel protein 1BL8-A (31) and the clathrin self-assembly (1B89-A) domain (32). The highest structural similarity found for AS-48 (Zscore 2.1, RMSD 3.6 Å) corresponds to NK-lysin, a 78-residue polypeptide with antibacterial and tumorolytic properties isolated from porcine small intestine (12). The solution structure of NK-lysin comprises also five amphipathic helices folded into a single globular domain (33). In Fig. 3, the three-dimensional structures of AS-48 and NK-lysin are compared. As may be seen in the figure, the relative position of helices α2 and α3 are very close in the two proteins, and so are, although to a less extent, helices α1 and α4. The largest difference is found for helix α5, which, interestingly, is located in the region where the C and N termini of NK-lysin meet. If one imagines the formation of a link between helices α5 and α1 to form a cyclic polypeptide, the similarity between the two proteins is greatly increased.
Electrostatic Properties of Bacteriocin AS-48.
Bacteriocin AS-48 has 14 charged residues—8 Lys, 2 Arg, and 4 Glu—which make it strongly positive charged, with an evaluated pI ≈ 10.5. This exceptionally high net positive charge becomes enhanced at pH 3, where all 4 Glu side chains are presumably protonated. Clusters of positive charges have been claimed to be responsible for the deleterious effects of plant and mammal defensins as well as of insect toxins on negatively charged cell membranes.
The amphipathic character of the five helices of AS-48 is not very large. The mean helical hydrophobic moments, 〈μH〉, as defined by Eisenberg and colleagues (34, 35), for helices 1 to 5 are 0.21, 0.22, 0.09, 0.22, and 0.13, respectively. These values are to be compared with a 〈μH〉 ≈ 0.70 for a helix of maximum amphiphilicity (Arg-Arg-Ile-Ile)3 spanning 12 residues. Only helices 1, 2, and 4 show a modest amphipathic character. On the contrary, α-helices in NK-lysin show a more marked amphipathic character, and the 〈μH〉 values for the corresponding helices are 0.41, 0.28, 0.45, 0.38, and 0.06.
Even so, it must be noted the highly asymmetrical distribution of the positive charge in AS-48, which can be noticed in the sequence, where all 10 basic residues are contained in a segment of 26 residues out of a total of 70. The charge distribution on the surface of the polypeptide was computed with the program grasp (36) for pH 7.0. Bacteriocin AS-48 has no His or Asp residues. Arg and Lys residues were taken as protonated and Glu residues as deprotonated. The electrostatic potential contoured at a level of 2kT/q is shown in Fig. 4, where it may be seen how the high positive potential rather than being evenly distributed all over the globular surface extends more largely in a region comprising helix α4 and the turn joining α4 and α5.
It is interesting to note that not all hydrophobic residues are buried at the core of the structure, but rather a significant number of them are exposed to the solvent. Fig. 5 shows two different faces of the polypeptide, front and back views generated by a 180° rotation around the z axis contained in the paper, in which the corresponding hydrophobic surfaces have been distinctly colored, and where their remarkable extension may be observed.
Figure 5.

Front view (same orientation as in Fig. 4) and back view (generated by a rotation of 180° around the z axis) showing in cyan the hydrophobic regions at the surface of bacteriocin AS-48.
Discussion
Knowledge of the three-dimensional structure in solution of bioactive substances with antibacterial properties interacting with biological cell membranes is a necessary requirement for the interpretation of their mode of action at the molecular level. We have determined herein the three-dimensional structure in aqueous solution of the bacteriocin AS-48 by NMR methods. It is noteworthy that AS-48 has the unusual characteristic of being a cyclic polypeptide. Interestingly, the head-to-tail union between Trp-70 and Met-1 lies in the middle of helix 5, which is shown to have a pronounced effect on the stability of the three-dimensional structure. Thus, preliminary results of the overexpression of the gene encoding AS-48 by recombinant methods show that the linear peptide is unable to form the native structure adopted by the cyclic polypeptide. Helix 5, spanning residues 64–5, and more specifically, the interaction of its internal hydrophobic residues (Val-67, Met-1, and Phe-5) with those in the internal core appears to be essential for the stability of the five-helical globule.
In the search for similar structures, two main families sharing significant structural similarities with bacteriocin AS-48 have been found. These were on one hand membrane-interactive proteins forming helical domain repeats, and on the other hand those related to the so-called saposin fold (33), which includes the effector polypeptide from porcine lymphocytes NK-lysin. We will concentrate on the latter ones because of a larger similarity of their structural features and biological properties to those of AS-48.
The “saposin fold” refers to the one experimentally determined by NMR for NK-lysin (33), considered to be the first member of the family, consisting of five α-helices folded into a globular domain (see Fig. 3), with a hydrophobic core and a hydrophilic surface. Other members in the family present a significant sequence homology with NK-lysin, including three conserved disulfide bridges and a set of hydrophobic residues. Among them we find the saposin polypeptides, which solubilize lipids and activate lysosomal hydrolases, the pulmonary surfactant-associated polypeptide SP-B, the pore-forming amoebapores, and parts of acid sphingomyelinase and acyloxyacylhydrolase (12). Furthermore, an interesting sequence homology has been detected in some plant aspartic proteinases, where the saposin motif appears to be circularly permuted (37). All these saposin-like proteins must adopt most likely the representative structure of NK-lysin on grounds of their conserved disulfide bridges and buried side chains, as well as the fact that insertions and deletions are mostly located in the loops joining the helices. Differences in surface residues and charge distribution must be at the origin of the diversity of functions among the different members of the family. All saposin-like polypeptides are highly resistant to thermal denaturation—i.e., they conserve their biological activities after incubation at 90–100°C—but these properties are abolished after reduction of the disulfide bonds (12, 38).
Even when there is no significant sequence homology between the two polypeptides, the three-dimensional structure of bacteriocin AS-48 can be fitted into the saposin fold with a minimum RMSD of 3.6 Å, once the backbone residues more alike have been selected and the most diverging ones have been removed (55 residues in a total of 70 were taken into consideration). This is in any case a relatively high score, which suggests a common antibacterial action. Both NK-lysin and AS-48 are extremely stable polypeptides, a guarantee that the structure is conserved after the interaction with their target cell membranes. The high stability of NK-lysin can be easily rationalized on terms of its three disulfide bridges and the amphipathic character of 3 of its 5 helices. It is not so obvious to justify the pronounced heat stability of AS-48, whose thermal denaturation temperature is 93°C (8). The head-to-tail additional peptide bond certainly contributes to it, and it may be complemented by an especially efficient interdigitation of hydrophobic side chains at the core of the globular structure, as manifested by the large number of contacts existing among them.
It is intriguing that there is close similarity in the topology of the two polypeptides when there is no sequence homology between them, and AS-48 lacks the key structural feature of the pattern of three disulfide bonds. When considering their biological action it is crucial to examine their electrostatic properties. Like AS-48, NK-lysin has a net 6 positive charges, most of which are arranged in a equatorial belt around the molecule, referring to a vertical position of helix 1 (39). The highest values of the calculated positive electrostatic potential are in the vicinity of helix 3, more specifically near its N terminus, although the value of the potential is high all along that helix. The positive electrostatic potential for AS-48 presents the highest values in the region comprising α4, the turn joining α4 and α5, and the N terminus of α5. Therefore there is not a correspondence between the location of the regions of maximum positive electrostatic potential in the structures of NK-lysin and AS-48. This is important because it would mean that the initial approach of the two polypeptides to a negatively charged membrane would be different in their orientation, because that approach will be reasonably guided by the direction of the maximum positive electrostatic potential.
Permeation of the cell membrane leading to cell death is the accepted mechanism for the action of a large number of membrane-lytic polypeptides. Antibacterial peptides containing high net positive charge, such as AS-48 and NK-lysin, show a preferential activity against bacteria and not normal mammalian cells, because their membranes are rich in acidic phospholipids (40). In previous studies (6), we have shown that bacteriocin AS-48 does interact with the cytoplasmic membrane of sensitive bacteria, inducing ion permeation, accompanied by the collapse of the membrane potential. More recently, a unifying concept, referred to as molecular electroporation, for the description of membrane pore formation by positively charged antibacterial peptides has been presented and exemplified by NK-lysin (39). Electroporation is the formation of pores in membranes attributable to the presence of an external electric field, and “molecular electroporation” denotes electroporation caused by the electric field produced by the binding of a charged molecule to the surface of a membrane. Molecular electroporation is expected if a reagent can bind to a lipid bilayer and it carries a sufficient charge density to provide an electrostatic potential of at least 0.2 V across the bilayer. This is shown to be like that from calculations of the electrostatic potential of NK-lysin associated with a membrane. Given that the net positive charge in AS-48 is as large as in NK-lysin, and that the charge appears to be more concentrated in a particular region of the molecule, it is to be expected that the conditions for molecular electroporation will also apply in the case of bacteriocin AS-48. According to that simple model, AS-48 initially would bind onto the surface of the target membrane; the peptide monomers would align themselves so that their hydrophilic surface would face the negatively charged phospholipid head groups; most probably, the three-dimensional structure of the polypeptide would be conserved on grounds of its pronounced stability; and polypeptide monomers should become associated at the surface by interaction of their hydrophobic surfaces, thus increasing the local concentration, favoring molecular electroporation and membrane permeation. Molecular electroporation could provide a mechanism for disorganization of the outer membrane of Gram-negative bacteria, facilitating the activity of AS-48 by a self-promoted uptake pathway. The increasing conductance of artificial membranes together with the overall membrane disorganization and formation of bleb-like protrusions observed on bacteria cells and liposomes treated with AS-48 (6, 41) also support this model.
Acknowledgments
We thank Mrs. C López, Mr. A. Gómez, and Mr. L. De la Vega for technical assistance, and Dr. G. Rivas for his help in the collection and analysis of data from the analytical ultracentrifuge. This work was supported by the Spanish Dirección General de Investigación Científica y Técnica, Project PB98-0677, and the Comisión Interministerial de Ciencia y Tecnología, Project BIO98-0908-CO2-01.
Abbreviations
- NOE
nuclear Overhauser enhancement
- RMSD
root mean square deviation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1e68).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210301097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210301097
References
- 1.Gálvez A, Maqueda M, Martínez-Bueno M, Valdivia E. Res Microbiol. 1989;140:57–68. doi: 10.1016/0923-2508(89)90060-0. [DOI] [PubMed] [Google Scholar]
- 2.Gálvez A, Valdivia E, Martínez M, Maqueda M. Can J Microbiol. 1989;35:318–321. doi: 10.1139/m89-048. [DOI] [PubMed] [Google Scholar]
- 3.Abriouel H, Valdivia E, Gálvez A, Maqueda M. Appl Environ Microbiol. 1998;64:4623–4626. doi: 10.1128/aem.64.11.4623-4626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez-Bueno M, Gálvez A, Valdivia E, Maqueda M. J Bacteriol. 1990;172:2817–2818. doi: 10.1128/jb.172.5.2817-2818.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Bueno M, Valdivia E, Gálvez A, Coyette J, Maqueda M. Mol Microbiol. 1998;27:347–358. doi: 10.1046/j.1365-2958.1998.00682.x. [DOI] [PubMed] [Google Scholar]
- 6.Gálvez A, Maqueda M, Martínez-Bueno M, Valdivia E. J Bacteriol. 1991;173:886–892. doi: 10.1128/jb.173.2.886-892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shai Y. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 8.Gálvez A, Giménez-Gallego G, Maqueda M, Valdivia E. Agents Chemother. 1989;33:437–441. doi: 10.1128/aac.33.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-Bueno M, Maqueda M, Gálvez A, Samyn B, van Beeumen J, Coyette J, Valdivia E. J Bacteriol. 1994;176:6334–6339. doi: 10.1128/jb.176.20.6334-6339.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samyn B, Martinez-Bueno M, Devreese B, Maqueda M, Gálvez A, Valdivia E, Coyette J, van Beeumen J. FEBS Lett. 1994;352:87–90. doi: 10.1016/0014-5793(94)00925-2. [DOI] [PubMed] [Google Scholar]
- 11.Langdon G M, Bruix M, Gálvez A, Valdivia E, Maqueda M, Rico M. J Biomol NMR. 1998;12:173–175. doi: 10.1023/a:1008267725043. [DOI] [PubMed] [Google Scholar]
- 12.Andersson M, Curstedt T, Jörnvall H, Johansson J. FEBS Lett. 1995;362:328–332. doi: 10.1016/0014-5793(95)00268-e. [DOI] [PubMed] [Google Scholar]
- 13.Laue T M, Shak B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 14.Kim H, Deonier R C, Williams J W. Chem Rev. 1997;77:659–690. [Google Scholar]
- 15.Ralston G. Introduction to Analytical Ultracentrifugation. Fullerton, CA: Beckman Instruments; 1993. [Google Scholar]
- 16.Hsu C S, Minton A P. J Mol Recognit. 1991;4:93–104. doi: 10.1002/jmr.300040208. [DOI] [PubMed] [Google Scholar]
- 17.Marion D, Wüthrich K. Biochem Biophys Res Commun. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 18.Aue W P, Bartholdi E, Ernst R R. J Chem Phys. 1976;64:2229–2246. [Google Scholar]
- 19.Bax A, Davis D G. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 20.Kumar A, Ernst R R, Wüthrich K. Biochem Biophys Res Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 21.Piotto M, Saudek V, Sklenár V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 22.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 23.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 24.Güntert P, Braun W, Wüthrich K. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 25.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 26.Van Gusteren W F, Berendsen H J C. Groningen Molecular Simulation (GROMOS) Library Manual. Groningen, The Netherlands: Biomos; 1987. [Google Scholar]
- 27.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 29.Richards F M. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Sander C. J Mol Biol. 1992;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 31.Doyle D A, Morais Cabral J, Pfuetzner R A, Kuo A, Gulbis J M, Cohen S L, Chait B T, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 32.Ybe J A, Brodsky F M, Hofmann K, Lin K, Liu S-H, Chen L, Earnest T N, Fletterick R J, Hwang P K. Nature (London) 1999;399:371–375. doi: 10.1038/20708. [DOI] [PubMed] [Google Scholar]
- 33.Liepinsh E, Andersson M, Ruysschaert J M, Otting G. Nat Struct Biol. 1997;4:793–795. doi: 10.1038/nsb1097-793. [DOI] [PubMed] [Google Scholar]
- 34.Eisenberg D, Weiss R M, Terwilliger T C. Nature (London) 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 35.Eisenberg D, Weiss R M, Terwilliger T C. Proc Natl Acad Sci USA. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholls A. grasp, Graphical Representation and Analysis of Surface Properties. New York: Columbia Univ.; 1993. [Google Scholar]
- 37.Guruprasad K, Tormakangas K, Kervinen J, Blundell T L. FEBS Lett. 1994;352:131–136. doi: 10.1016/0014-5793(94)00935-x. [DOI] [PubMed] [Google Scholar]
- 38.Leippe M, Tannich E, Nickel R, van der Goot G, Pattus F, Horstmann R D, Müller-Eberhard H J. EMBO J. 1992;11:3501–3506. doi: 10.1002/j.1460-2075.1992.tb05432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miteva M, Andersson M, Karshikoff A, Otting G. FEBS Lett. 1999;462:155–158. doi: 10.1016/s0014-5793(99)01520-3. [DOI] [PubMed] [Google Scholar]
- 40.Brock T D. Biology of Microorganisms. 2nd Ed. Englewood Cliffs, NJ: Prentice Hall; 1974. [Google Scholar]
- 41.Mendoza F, Maqueda M, Gálvez A, Martínez-Bueno M, Valdivia E. Appl Environ Microbiol. 1999;65:618–625. doi: 10.1128/aem.65.2.618-625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]