Abstract
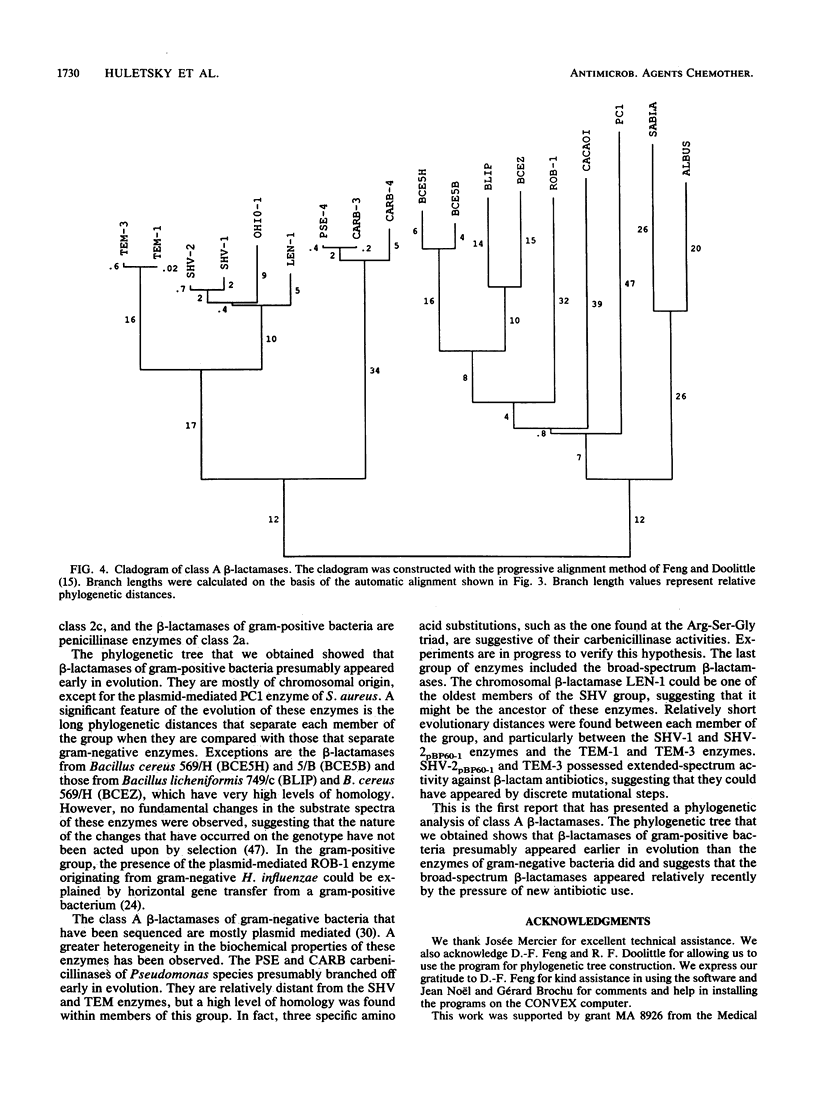
We determined the nucleotide sequence of the blaSHV-2(pBP60-1) gene from Klebsiella ozaenae which confers resistance to broad-spectrum cephalosporins. The structural gene encodes a polypeptide product of 286 amino acids, and the estimated molecular weight of the mature protein is 28,900. Amino acid sequence comparison of the SHV-2pBP60-1 enzyme with all known class A beta-lactamases and homology studies showed that the residues were highly conserved. Furthermore, SHV-2pBP60-1 was clearly related to SHV-1, LEN-1, and OHIO-1. The SHV-2pBP60-1 enzyme differed from SHV-1 isolated from Klebsiella pneumoniae by seven amino acid substitutions. One of these substitutions, the Gly----Ser substitution at position 234, is probably a key region for the novel activity of cefotaxime hydrolysis. A phylogenetic tree was constructed by using all class A beta-lactamases of known sequences by a progressive alignment method. The data suggested that the beta-lactamases of gram-positive Streptomyces, Staphylococcus, and Bacillus species appeared early in evolution, followed by the PSE and CARB enzymes of Pseudomonas species and, more recently, by the SHV-type and TEM-type enzymes found in enteric bacteria. Larger evolutionary distances separated clusters of the gram-positive beta-lactamases than separated clusters of the gram-negative enzymes. Results of this phylogenetic study suggested that extended-spectrum enzymes are recent derivatives that are selected by the use of new cephalosporins.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Arakawa Y., Ohta M., Kido N., Fujii Y., Komatsu T., Kato N. Close evolutionary relationship between the chromosomally encoded beta-lactamase gene of Klebsiella pneumoniae and the TEM beta-lactamase gene mediated by R plasmids. FEBS Lett. 1986 Oct 20;207(1):69–74. doi: 10.1016/0014-5793(86)80014-x. [DOI] [PubMed] [Google Scholar]
- Barthélémy M., Peduzzi J., Labia R. Complete amino acid sequence of p453-plasmid-mediated PIT-2 beta-lactamase (SHV-1). Biochem J. 1988 Apr 1;251(1):73–79. doi: 10.1042/bj2510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélémy M., Péduzzi J., Ben Yaghlane H., Labia R. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 1988 Apr 11;231(1):217–220. doi: 10.1016/0014-5793(88)80734-8. [DOI] [PubMed] [Google Scholar]
- Boissinot M., Levesque R. C. Nucleotide sequence of the PSE-4 carbenicillinase gene and correlations with the Staphylococcus aureus PC1 beta-lactamase crystal structure. J Biol Chem. 1990 Jan 15;265(2):1225–1230. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Buré A., Legrand P., Arlet G., Jarlier V., Paul G., Philippon A. Dissemination in five French hospitals of Klebsiella pneumoniae serotype K25 harbouring a new transferable enzymatic resistance to third generation cephalosporins and aztreonam. Eur J Clin Microbiol Infect Dis. 1988 Dec;7(6):780–782. doi: 10.1007/BF01975048. [DOI] [PubMed] [Google Scholar]
- Bush K. Characterization of beta-lactamases. Antimicrob Agents Chemother. 1989 Mar;33(3):259–263. doi: 10.1128/aac.33.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 1, 2a, 2b, and 2b'. Antimicrob Agents Chemother. 1989 Mar;33(3):264–270. doi: 10.1128/aac.33.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T. Nucleotide sequence of the Staphylococcus aureus PC1 beta-lactamase gene. Nucleic Acids Res. 1986 Jul 25;14(14):5940–5940. doi: 10.1093/nar/14.14.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehottay P., Dusart J., De Meester F., Joris B., Van Beeumen J., Erpicum T., Frère J. M., Ghuysen J. M. Nucleotide sequence of the gene encoding the Streptomyces albus G beta-lactamase precursor. Eur J Biochem. 1987 Jul 15;166(2):345–350. doi: 10.1111/j.1432-1033.1987.tb13521.x. [DOI] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann L., Ferré B., Goldstein F. W., Rizk N., Pinto-Schuster E., Acar J. F., Collatz E. SHV-5, a novel SHV-type beta-lactamase that hydrolyzes broad-spectrum cephalosporins and monobactams. Antimicrob Agents Chemother. 1989 Jun;33(6):951–956. doi: 10.1128/aac.33.6.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Hussain M., Pastor F. I., Lampen J. O. Cloning and sequencing of the blaZ gene encoding beta-lactamase III, a lipoprotein of Bacillus cereus 569/H. J Bacteriol. 1987 Feb;169(2):579–586. doi: 10.1128/jb.169.2.579-586.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarlier V., Nicolas M. H., Fournier G., Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988 Jul-Aug;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juteau J. M., Levesque R. C. Sequence analysis and evolutionary perspectives of ROB-1 beta-lactamase. Antimicrob Agents Chemother. 1990 Jul;34(7):1354–1359. doi: 10.1128/aac.34.7.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebe C., Nies B. A., Meyer J. F., Tolxdorff-Neutzling R. M., Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985 Aug;28(2):302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz L. C., Blanken R. L. A practical method for calculating evolutionary trees from sequence data. J Theor Biol. 1981 Jul 21;91(2):261–272. doi: 10.1016/0022-5193(81)90233-2. [DOI] [PubMed] [Google Scholar]
- Knothe H., Shah P., Krcmery V., Antal M., Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983 Nov-Dec;11(6):315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- Medeiros A. A. Beta-lactamases. Br Med Bull. 1984 Jan;40(1):18–27. doi: 10.1093/oxfordjournals.bmb.a071942. [DOI] [PubMed] [Google Scholar]
- Medeiros A. A., Levesque R., Jacoby G. A. An animal source for the ROB-1 beta-lactamase of Haemophilus influenzae type b. Antimicrob Agents Chemother. 1986 Feb;29(2):212–215. doi: 10.1128/aac.29.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier J., Levesque R. C. Cloning of SHV-2, OHIO-1, and OXA-6 beta-lactamases and cloning and sequencing of SHV-1 beta-lactamase. Antimicrob Agents Chemother. 1990 Aug;34(8):1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Neugebauer K., Sprengel R., Schaller H. Penicillinase from Bacillus licheniformis: nucleotide sequence of the gene and implications for the biosynthesis of a secretory protein in a Gram-positive bacterium. Nucleic Acids Res. 1981 Jun 11;9(11):2577–2588. doi: 10.1093/nar/9.11.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Philippon A., Labia R., Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989 Aug;33(8):1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M., Currie-McCumber C., Hull A., Behlau I., Kron M. OHIO-1 beta-lactamase is part of the SHV-1 family. Antimicrob Agents Chemother. 1990 Aug;34(8):1570–1576. doi: 10.1128/aac.34.8.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M., Medeiros A. A., Kron M. A., Currie-McCumber C., Papa E., Vartian C. V. Novel plasmid-mediated beta-lactamase in members of the family Enterobacteriaceae from Ohio. Antimicrob Agents Chemother. 1986 Aug;30(2):220–224. doi: 10.1128/aac.30.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloma A., Gross M. Molecular cloning and nucleotide sequence of the type I beta-lactamase gene from Bacillus cereus. Nucleic Acids Res. 1983 Jul 25;11(14):4997–5004. doi: 10.1093/nar/11.14.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


