Abstract
By comparing 4,344 protein sequences from fission yeast Schizosaccharomyces pombe with all available eukaryotic sequences, we identified those genes that are conserved in S. pombe and nonfungal eukaryotes but are missing or highly diverged in the baker's yeast Saccharomyces cerevisiae. Since the radiation from the common ancestor with S. pombe, S. cerevisiae appears to have lost about 300 genes, and about 300 more genes have diverged by far beyond expectation. The most notable feature of the set of genes lost in S. cerevisiae is the coelimination of functionally connected groups of proteins, such as the signalosome and the spliceosome components. We predict similar coelimination of the components of the posttranscriptional gene-silencing system that includes the recently identified RNA-dependent RNA polymerase. Because one of the functions of posttranscriptional silencing appears to be “taming” of retrotransposons, the loss of this system in yeast could have triggered massive retrotransposition, resulting in elimination of introns and subsequent loss of spliceosome components that become dispensable. As the genome database grows, systematic analysis of coordinated gene loss may become a general approach for predicting new components of functional systems or even defining previously unknown functional complexes.
A major outcome of the recent advances in comparative genomics is the realization of the major role of horizontal gene transfer and lineage-specific gene loss in the evolution of prokaryotes (1–4). These phenomena appear to account largely for the remarkable diversity of the prokaryotic gene repertoires. The likelihood of horizontal gene transfer between eukaryotes, at least multicellular ones, is low because, for a gene to be laterally transferred, it must enter the germ line. In contrast, there is no such restriction for gene loss. The dramatic variation in the number of genes among eukaryotes, in some cases even between rather closely related species—yeast Saccharomyces cerevisiae, for example, has about 6,000 genes compared with at least 8,000–9,000 in multicellular ascomycetes such as Aspergillus (5)—suggests that, along with proliferation of gene families (6), lineage-specific gene loss could have been important in eukaryotic evolution.
The two yeasts, S. cerevisiae and Schizosaccharomyces pombe, are probably the optimal current choice of genomes to compare with the aim of estimating the number of lost genes. The genome of S. cerevisiae, arguably the best-studied eukaryote in terms of gene functions (6), has been completed (7), and for S. pombe, up to 70% of the genome sequence is available. For estimating gene loss, it is critical to have assurance that (nearly) all genes in the analyzed genome have been identified; S. cerevisiae is the only eukaryotic genome for which such confidence exists, particularly because of the paucity of introns, which facilitates gene detection. The two yeast species are close enough so that direct counterparts among their genes [orthologs (8)] are readily identifiable but distant enough so that differences among their gene repertoires are substantial. By comparing 4,344 available protein sequences from fission yeast S. pombe with all eukaryotic sequences, we identified those genes that are conserved in S. pombe and nonfungal eukaryotes but are missing or highly diverged in the baker's yeast S. cerevisiae. We describe patterns of coelimination of functionally linked genes that can be used for predicting new components of known complexes and pathways as well as new functional systems.
Materials and Methods
The database searches and other sequence manipulations were carried out by using programs of the SEALS package (9). Searches of the Nonredundant protein sequence database at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD) were performed by using the gapped blastp program and, for in-depth analysis, the iterative psi-blast program (10). Profile searches for protein domains were conducted by using a previously constructed and updated library of position-specific score matrices (PSSMs; ref. 5 and L.A., unpublished data) generated by using the psi-blast program, which typically was run with the cut-off of E = 0.01 for inclusion of sequences into the PSSM, or by using the SMART tool (11). Classification of yeast proteins by functional categories was based on previously published data, the results of domain-specific profile searches (6), and the clusters of orthologous groups of proteins (COGs) database (12).
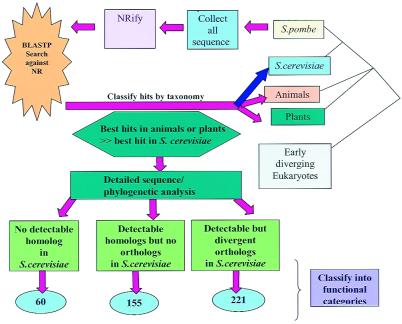
The protocol used to identify candidates for gene loss and divergence is outlined in Fig. 1. The phyletic distribution of the significant database hits (expectation value, E < 0.001 in a gapped blast search) in the nonredundant database for a nonredundant set of 4,344 S. pombe proteins was analyzed by using the tax_collector program of the SEALS package. The initial set of S. pombe proteins that could have been lost or have diverged beyond expectation in S. cerevisiae was compiled of those proteins that registered a statistically significant hit in at least one animal or plant species, but not in S. cerevisiae, and those that hit an animal or plant sequence with an E value at least 10 orders of magnitude lower (more significant) than that of the best hit to a S. cerevisiae sequence. This preliminary detection of possible gene losses was followed with manual evaluation of each candidate, which included psi-blast searches to detect potential diverged counterparts in S. cerevisiae, and phylogenetic analysis for cases when two or more members of a paralogous family were present in S. pombe as opposed to just one member in S. cerevisiae.
Figure 1.
The protocol for detecting candidates for gene loss and divergence in S. cerevisiae. nrify is a program in the SEALS package that is used to generate a nonredundant protein database.
Multiple alignments were constructed by using the clustalw program (13) and adjusted manually based on the outputs of psi-blast searches and structural considerations. Distance matrices were constructed from the alignments by using the protdist program of the PHYLIP package, which employs the Dayhoff's PAM 001 matrix for the calculation of evolutionary distances (14). Phylogenetic trees were generated by using the neighbor-joining method as implemented in the neighbor program of the PHYLIP package, with 1,000 bootstrap replications used to assess the reliability of each node (14).
Results
Three Classes of Genes Apparently Lost or Highly Diverged in S. cerevisiae.
Under the protocol shown in Fig. 1, a nonredundant set of S. pombe protein sequences was compared with the complete protein database, and statistically significant hits to proteins from S. cerevisiae, animals, plants, and other eukaryotes were recorded separately. The majority of S. pombe protein sequences are expected to be significantly more similar to their orthologs from S. cerevisiae compared with those from any species outside fungi, and this is the clear trend observed when the database hits are classified by their taxonomic origin (Fig. 2). Whenever an anomaly is seen, i.e., a S. pombe protein is more similar to a homolog from another eukaryotic lineage compared with those from S. cerevisiae, there seems to be a potential case of gene loss or rapid divergence in the S. cerevisiae lineage (Figs. 1 and 2). The S. pombe proteins that showed such unexpected behavior (Fig. 2) were subjected to case-by-case validation, which involved iterative database searches and phylogenetic analysis, to identify potential diverged homologs in S. cerevisiae and to clarify the issue of orthology (Fig. 1 and Materials and Methods).
Figure 2.
Scatter plots of sequence similarity between S. pombe proteins and their homologs from S. cerevisiae, plants, and animals. Horizontal axis: −log10[E value (S. pombe with S. cerevisiae)]. Vertical axis: log10[E-value (S. pombe with S. cerevisiae)] − log10 [E-value (S. pombe with nonfungal eukaryote)]. The thick, horizontal line indicates the 10 orders of magnitude threshold used to delineate the set of anomalous hits that were analyzed further for likely cases of gene loss and divergence. (A) Metazoa vs. S. cerevisiae. (B) Plants vs. S. cerevisiae
After some of the anomalous hits detected at the first, automatic stage (Fig. 1) were discarded as inconclusive, the remaining cases were classified into three categories: (i) S. pombe proteins with no detectable homologs in S. cerevisiae; (ii) S. pombe proteins that had paralogs in S. cerevisiae, but no apparent orthologs; and (iii) S. pombe proteins that appeared to have highly diverged orthologs in S. cerevisiae.
The proteins in the first two categories are likely to represent genes that have been present in the common ancestor of animals, plants, and fungi, but have been lost (or have diverged beyond recognition) in the S. cerevisiae lineage. For category (ii), reciprocal database searches with the respective S. cerevisiae proteins as queries indicated the existence of distinct orthologs in S. pombe, different from the proteins that showed anomalous hits. Phylogenetic tree analysis and/or comparison of domain architectures showed that the latter had no counterparts in S. cerevisiae. In phylogenetic trees, these S. pombe genes typically formed coherent orthologous groups with their counterparts from animals and/or plants; in contrast, in the sister clusters from the same family of paralogs, the S. cerevisiae and S. pombe proteins grouped together as expected (Fig. 3). These are potential cases of partial functional redundancy in which S. cerevisiae apparently has lost one or more members of an ancestral paralogous family. Altogether, this analysis, performed by using ≈70% of S. pombe genes, suggested that S. cerevisiae had lost at least 200 genes, or about 3% of its gene complement, since its radiation from the common ancestor with S. pombe, with about the same number of genes having diverged at unexpectedly high rates (Fig. 1).
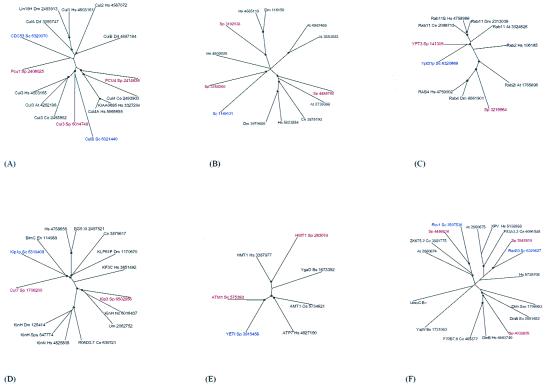
Figure 3.
Selected phylogenetic trees for protein families with apparent loss of paralogs in S. cerevisiae. (A) Cullins. (B) Cyclins (two cases of gene loss in S. cerevisiae). (C) Rab-type GTPase. (D) Kinesin ATPase subunit. (E) ABC transporter ATPase subunit. (F) DinB family of DNA repair polymerases. Circles denote nodes with at least 70% bootstrap support. The proteins are designated by their gene identifiers (GI numbers) in the nonredundant database and species abbreviations: Sc, S. cerevisiae; Sp, S. pombe; Nc, Neurospora crassa; Um, Ustilago maydis; Dm, D. melanogaster; Ce, C. elegans; Hs, Homo sapiens; Xl, Xenopus laevis; At, Arabidopsis thaliana; Os, Oryza sativa. Proteins from S. cerevisiae are color-coded blue, and those from S. pombe are coded red.
Coelimination and Codivergence of Functionally Linked Genes.
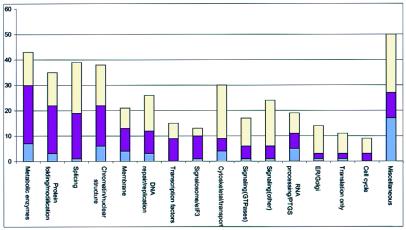
Apparent gene loss and divergence in S. cerevisiae cover the entire spectrum of cellular functions, including basic ones, such as translation (Fig. 4). On excluding the broad classes, such as miscellaneous proteins and metabolic enzymes, it is apparent that a large fraction of the gene loss and a significant amount of divergence are seen in functionally well defined groups of proteins that are involved in nuclear structure maintenance, pre-mRNA splicing, RNA modification, posttranscriptional gene silencing (PTGS), and protein folding/processing (Fig. 4).
Figure 4.
The extent of apparent gene loss in different functional categories of S. cerevisiae genes. Three types of evolutionary events, namely, complete elimination of a gene family, with no detectable S. cerevisiae homolog for a S. pombe protein (Bottom, purple area in each bar), elimination of a paralog (Middle, dark-red area), and anomalous divergence (Top, yellow area) are shown for different functional classes of S. cerevisiae genes. Vertical axis, number of genes. ER, endoplasmic reticulum.
For most functional groups, only a small fraction of the total number of proteins has been lost. However, several distinct pathways and complexes stand in sharp contrast to this trend, suggesting coelimination of functionally interacting sets of proteins (Table 1). A clear-cut example is the eIF3/signalosome complex that participates in multiple protein–protein interactions mediating signaling, translation, and protein degradation (15, 16). Although the majority of these proteins are conserved in animals, plants, and S. pombe (17), most of them seem to be missing in S. cerevisiae, indicating that they probably have been lost as a group (Table 1). This leads to the prediction that the principal signalosome function does not exist in S. cerevisiae. Similarly, the ortholog of the repair protein XP-E is conserved throughout the crown group of eukaryotes, but is missing in S. cerevisiae, along with the Cullin 4A ortholog. Recently, these proteins have been shown to interact physically (18), which implies a functional interaction and a concomitant loss of the respective genes in the S. cerevisiae lineage. Thus, it appears that examination of the patterns of gene loss, along with a careful analysis of the conserved domains that serve as functional signatures, may help in reconstructing potential functional interactions and perhaps predicting unknown pathways and complexes. A few such cases of potential functional grouping of experimentally uncharacterized proteins are discussed here (Table 1).
Table 1.
Coelimination and codivergence of functionally linked genes in S. cerevisiae*
| Protein function and representative in S. pombe† | Domain architecture‡ | L/D§ | Comments |
|---|---|---|---|
| Posttranscriptional gene
silencing/mRNA stability and modification | |||
| RNA-dependent RNA polymerase (3169081) | RNA-dependent RNA polymerase | L | Involved in posttranscriptional silencing in plants and Neurospora |
| Argonaute/eIF2C (2330856) | Argonaute | L | Involved in RNA I-mediated silencing in C. elegans |
| RNA helicase-nuclease (1351642) | SFII-Helicase+2*RNASEIII | L | Ortholog of plant protein carpel factory involved in regulation of cell proliferation |
| RNAse PH family protein (1723274) | RNAse PH | L | |
| 3′ → 5′ exonuclease (4007797) | 3′ → 5′ exonuclease | L | Ortholog of the deadenylating nuclease involved in RNA decay |
| Inactive RNA helicase (4160338) | SFI-helicase | L | Ortholog of the animal protein Aquarius, a superfamily I helicase with disrupted catalytic motifs (L.A. and E.V.K., unpublished data) |
| RNA helicase (6048290) | SFI-helicase | L | |
| Zn-knuckle RNA-binding protein (3738189) | 5*Zn-knuckle | L | A paralog of the clipper/polyadenylation complex subunit CPSF30 |
| NMD2 (2388907) | 2*NIC domain | D | An adapter protein in the nonsense-mediated RNA decay pathway¶ |
| NAM7 helicase (3581879) | SFI-helicase | D | Helicase involved in nonsense-mediated RNA decay¶ |
| Spliceosome | |||
| U2AF, 23-kDa subunit (6136086) | Zn-knuckle+RRM+Zn-knuckle | L | RNA-binding protein of the U2 snRNP |
| U2AF, 59 kDa (549144) | RRM | L | RNA-binding protein of the U2 snRNP |
| U5 snRNP, 40-kDa subunit (4495124) | WD40 protein | L | Predicted adapter-mediating protein–protein interactions |
| U2 B" protein (3169094) | 2RRM* | L | Predicted RNA-binding protein |
| Component of U1 RNP Yhc1p (3006184) | C2H2 finger domain | D | Predicted RNA-binding protein |
| PRP21 ortholog (2414602) | SWAP domain | D | Along with PRP9, activates U2 snRNP to bind pre-mRNA |
| PRP9 ortholog (3135996) | C2H2 finger domain | D | Predicted RNA-binding protein |
| PRP39 (3169096) | TPR repeats | D | Predicted adapter mediating protein–protein interactions |
| Predicted RNA-binding protein (2104448) | C4+C2H2+RRM+G-patch | L | Multiple RNA-binding domains |
| Predicted RNA-binding protein (2879872) | RRM+PWI | L | Predicted RNA binding with spliceosome-associated PWI domain |
| Predicted RNA-binding protein (3810835) | NTF2+RRM | L | Could be involved in the transport of pre-mRNA base on the NTF2 domain |
| Predicted RNA-binding protein (1351664) | 2*KH | L | |
| Predicted RNA-binding protein (6066740) | PWI | L | PWI domain is present in several splicing factors and could bind RNA |
| Predicted RNA-binding protein (2467274) | RRM | D | |
| Chromatin/nuclear structure-associated proteins | |||
| Clr4 (3334847) | Chromodomain+SET | L | Predicted methyltransferase involved in transcriptional silencing |
| SWI6 (730857) | Chromodomain | L | Heterochromatinic chromodomain protein ortholog |
| Inactivated cytosine-specific DNA methyltransferase (730347) | SAM-binding methylase domain | L | Apparently inactive ortholog of animal silencing-associated methylases of animals |
| MLO2 (2498563) | PHD finger | L | Affects chromosome segregation when overexpressed |
| BAF53 homolog (1351610) | Actin | L | SNF/SWI complex-associated Actin |
| NASP ortholog (5830515) | NASP domain | L | Member of a highly conserved family of Histone-binding proteins |
| Nuclear matrix protein p84 ortholog (6562903) | Unique domain | L | The vertebrate ortholog binds Retinoblastoma protein |
| ASH2 ortholog (3080533) | SPRY+PHD | L | Orthologous Drosophila protein ASH2 is a part of the chromatin-associate Trithorax complex |
| Predicted chromatin-associated, DNA-binding protein (5706512) | PHD+JOR+PHD | L | Homolog of human XE169 protein; the JOR domain may possess an as yet unknown enzymatic activity |
| Predicted chromatin protein (5441491) | SWI3+Rossmann-fold oxidase +HMG1 | L | Novel configuration of domains; may be a hitherto unexplored, chromatin-associated, NAD/FAD-dependent enzyme |
| Predicted adenine-specific DNA methylase (1175468) | SAM-binding methylase domain | L | Belongs to the Kar4/Ime4 family of adenine methylases; could be involved in a novel pathway of transcription/chromatin structure regulation |
| Predicted chromatin protein (2370493) | JOR | L | May possess an as yet unknown enzymatic activity |
| Predicted chromatin protein (1351640) | SKI/SNW | D | Homolog of the animal SKIP protein involved in transcriptional regulation |
| DNA repair/replication/damaged DNA-associated checkpoints | |||
| Hus1 (3219811) | PCNA clamp | L | Component of the damaged DNA-sensing complex |
| RAD9 (131816) | PCNA clamp | L | Component of the damaged DNA-sensing complex |
| RAD17 (1709996) | Clamp loader (AAA+) ATPase | D | Component of the damaged DNA-sensing complex |
| DINB1 ortholog (4038629) | DNA polymerase V+2HhH* | L | Translesion DNA repair polymerase |
| AlkB ortholog (3080529) | AlkB | L | Predicted hydrolytic enzyme involved in alkylated DNA repair |
| G/T mismatch-specific thymine DNA glycosylase (3915098) | DNA glycosylase | L | |
| Endonuclease V (1723511) | EndoV | L | Endonuclease involved in DNA repair |
| Telomerase (2340169) | Reverse transcriptase | D | Telomere maintenance |
| Ciliate-type telomere-binding protein (7491013) | OB-fold domain-containing single-stranded DNA-binding protein | L | Telomere maintenance |
| Replication protein A (2498845) | OB-fold | D | Replication initiation |
| CDC18 (1168808) | (AAA+) ATPase | D | S phase–mitosis coupling |
| ORC1 (1709487) | BAM+(AAA+) ATPase | D | ATPase subunit of the origin-recognition complex |
| ORC3 Latheo (6224782) | ORC3 | D | Origin recognition complex subunit |
| ORC5 (6093628) | Inactive AAA+ ATPase | D | ATP-binding but not hydrolyzing origin recognition complex subunit |
| TRF4/5 family protein (3219960) | Polβ family nucleotidyltransferase | L | Predicted to be involved in DNA repair in conjunction with topoisomerase I |
| Terminal Nucleotidyl transferase (1175369) | polβ family nucleotidyl transferase | L | Ortholog of vertebrate terminal deoxynucleotidyl transferases |
| Predicted DNAase (3417434) | SAP+3′ → 5′ nuclease (KapD family) | L | Predicted to localize to regions of active chromatin |
| Predicted A/G mismatch-specific DNA glycosylase (1723233) | DNA glycosylase+HhH | L | |
| Signalosome/eIF3/proteasome | |||
| EIF3 p66 (4056551) | Unique domain | L | |
| EIF3 p48/int6 (4160345) | PINT | L | Component of both signalosome and eIF3 |
| GPS1 ortholog (3873540) | PINT | L | Negative regulator of AP-1 transcription |
| EIF3 p40 (6014439) | PAD1/JAB1 | L | Component of both signalosome and eIF3 |
| EIF3 p167 ortholog (3650404) | Unique domain | D | eIF3 component |
| EIF3 Tif31p (6491837) | Unique domain | D | eIF3 component |
| Predicted signalosome subunit (5731945) | PINT | L | |
| Predicted signalosome subunit (2414596) | PINT | L | |
| Predicted signalosome subunit (3327876) | PINT | L | |
| Predicted signalosome subunit (2832888) | PAD1/JAB1 | L | |
| Protein folding, modification, and processing | |||
| BAG-2 homolog (3133105) | Ubiquitin | L | Ubiquitin-like lysine modification of proteins |
| Leucine aminopeptidase (1175415) | l-Aminopeptidase | L | Exoproteolytic processing of proteins |
| Peptidylprolyl isomerase (3169061) | WD40+cyclophilin | L | |
| Peptidylprolyl isomerase (1351676) | U-box+cyclophilin | L | A chaperone potentially involved in the assembly of proteosome-type complexes |
| Peptidylprolyl isomerase (5738526) | Cyclophilin+RRM | L | A chaperone potentially involved in the spliceosome assembly |
The cases in which experimental evidence for a role in the given pathway or complex exists for the particular S. pombe protein or its ortholog from another organism are shown in boldface. The remaining (not boldfaced) proteins in each category are predicted to belong to the same pathway or complex on the basis of coelimination and codivergence combined with analysis of domain composition.
GenBank identifiers (GI numbers) for the respective S. pombe proteins are given in parentheses.
Domains are shown consecutively from the N terminus to the C terminus. Known and predicted nucleic acid-binding domains: RRM (RNA recognition motif), Zn-knuckle, C4, C2H2 and PHD Zn-fingers, G-patch, HhH (helix–hairpin–helix), SAP (SAF-A/B, Acinus, and PIAS), SPRY (SplA ryanodine receptor domain), SWAP (suppressor of white apricot), OB (oligomer-binding)-fold, PWI (PWI motif-containing domain); protein–protein interaction adapter domains: WD40, TPR (tetratricopeptide) repeats, BAM (bromo-associated motif), NIC (NMD2, eIF4G, CBP80), PINT (proteasomal subunits, Int-6, Nip-1, Trip-15), PAD1/JAB1 (Jun activation domain binding), SKI/SNW (domain found in SKIP/SnwA proteins), PCNA (proliferating cell nuclear antigen), U (ubiquitination) box. Other, including enzymatic, domains include SFI, SFII [superfamily I and II (helicases)], JOR (Jumonji-related), AAA+ (a superfamily of ATPases including the classic AAA proteins), EndoV (endonuclease V domain), and NTF2 (nuclear transport factor 2 domain). For detailed information on most of these domains, see the SMART web site, http://smart.embl-heidelberg.de/.
L, apparent gene loss; D, gene divergence.
The correlation between the divergence of these genes with the loss of the PTGS suggests the possibility of a functional connection between the latter and nonsense-mediated mRNA decay.
Prediction of Previously Unknown Functional Interactions Using Coelimination and Codivergence Patterns.
Several known components of the spliceosome that functionally interact in other eukaryotes are missing in S. cerevisiae, and for several others, the S. cerevisiae orthologs are highly diverged, indicating that gene loss and divergence have extensively affected the yeast spliceosome (Table 1). Thus, proteins that have been lost or diverged in S. cerevisiae and contain domains compatible with a function in splicing are likely to be as yet unidentified spliceosome components. Several such proteins that are conserved in the eukaryotic crown group, but not in S. cerevisiae, and contain RNA-binding domains typically associated with splicing were detected (Table 1). The loss of spliceosomal components in S. cerevisiae correlates with the scarcity of spliceosomal introns (19). A likely evolutionary scenario involves the elimination of the majority of introns, probably through reverse transcription, followed by the loss of many specialized components of the splicing machinery. Many of the surviving spliceosomal components could have diverged rapidly to adapt to their new milieus.
Correlated loss is observed also among proteins associated with nonspliceosomal RNA-processing events such as poly(A)-tail addition and RNA modification and degradation. Recently, it has been shown that PTGS in plants, animals, and fungi is mediated by an RNA degradation system that includes an RNA-dependent RNA polymerase (RDRP) (20, 21), nucleases (22, 23), and a conserved protein of the Argonaute/eIF2C family (24). S. cerevisiae has lost both the RDRP, the critical enzyme of this pathway, and the Argonaute family protein that S. pombe shares with multicellular eukaryotes. This raises the possibility of other potential components of this pathway being among the genes lost in S. cerevisiae, and a notable assemblage of such candidates, including predicted helicases and nucleases, has been detected (Table 1). Carpel factory, the plant ortholog of the RNA helicase–RNase III protein (Table 1), has been proposed to negatively regulate floral meristem proliferation (25). The present observations suggest that it may function through posttranscriptional gene silencing, in conjunction with the other components of this pathway. Higher-order functional networks with the eIF3 subunits and the translation apparatus also might exist, given that the Argonaute-like proteins are possible translation regulators (eIF2C) (26). A direct evolutionary connection between the loss of PTGS components and spliceosome components is conceivable because it has been shown that PTGS significantly contributes to the control of transposition (24, 27). The loss of genes coding for PTGS components in the S. cerevisiae lineage could have induced a burst of retrotransposition (28), wiping out most of the intron-containing genes and leading to the subsequent deterioration of the spliceosome.
A pattern of coelimination is seen also among proteins involved in chromatin remodeling such as SWI6, CLR4, ASH2, and MLO2 (Fig. 4). Several other lost proteins can be identified as likely components of chromatin-associated regulatory complexes (Table 1). Notably, this subset of genes apparently lost by S. cerevisiae includes several methyltransferases, namely, a DNA-cytosine methylase, a predicted Kar4/Ime4-type DNA-adenine methylase, and two predicted methylases of chromatin proteins from the SET-domain superfamily (Table 1; L.A. and E.V.K., unpublished data). It seems likely that these methylases, together with other chromatin-associated proteins that are missing in S. cerevisiae, are involved in chromatin-level gene silencing that could interface, in a fashion yet unknown, with the PTGS. This is consistent with the recent evidence from plants that the RNA-mediated gene silencing could directly affect chromatin-level silencing through methylation of nuclear DNA (29, 30).
Among the genes involved in protein folding and modification, a striking feature is the loss, in S. cerevisiae, of three peptidyl-prolyl isomerases of the cyclophilin family, which have unusual domain architectures (Table 1). These chaperones could have been coeliminated with other functional complexes whose assembly they might have controlled. In particular, the cyclophilin-RRM protein could be involved in RNA-dependent spliceosome assembly.
Other gene losses with interesting functional implications are seen in DNA repair and replication pathways (Table 1). These include two proteins of the proliferating cell nuclear antigen/Pol IIIβ fold, Hus1 and Rad9, which are implicated in a damaged-DNA-dependent checkpoint (31, 32). It can be predicted that this particular checkpoint, that probably has been eliminated in S. cerevisiae, also involves XP-E and possibly some of the nucleases that have been lost in yeast (Table 1). Animals, S. pombe, plants, and ciliates all share a telomere-binding protein that contains OB-fold domains and interacts with the telomerase (33); this ancient eukaryotic gene has been lost in S. cerevisiae. The S. cerevisiae single-strand DNA-binding protein CDC13p (34), which has no detectable homologs in other species, appears to be a lineage-specific functional displacement for the ancestral telomere-binding proteins to form a unique telomeric structure. This is consistent with the drastic divergence of the S. cerevisiae telomerase reverse transcriptase subunit (Table 1).
Codivergence of functionally linked genes is as noticeable as coelimination (Table 1). In addition to the spliceosome components, a codivergent group is composed of proteins involved in initiation of replication, namely, the origin recognition complex (ORC) subunits (35). This divergence could reflect functional connections between ORC and chromatin components that have been lost or have diverged in yeast (Table 1).
Discussion
When extrapolated to the entire S. pombe genome, the above results indicate that at least 300 genes that have been present in the common ancestor of fungi, plants, and animals have been lost and another 300 or so have diverged far beyond expectation in the S. cerevisiae lineage. This estimated loss and divergence of about 10% of the S. cerevisiae gene complement is the lower bound of the extent of these phenomena because the protocol used here does not detect gene losses that occurred before the radiation of the two yeast lineages from their common ancestor. The notion of significant gene loss in fungi is consistent with the fact that microsporidia, probable early members of the fungal clade, possess extremely reduced gene complements (36).
We employed sequence comparison, using blast as a preliminary screen for proteins with anomalous evolutionary behavior, which was followed by a validation step including a search for potential weak similarities and phylogenetic analysis. Scores and E values reported by blast are measures of the similarity between protein sequences, and differences between them do not necessarily accurately reflect evolutionary relationships. Nevertheless, the use of such criteria, at least in first-pass, automatic screens, seems to be justified because they have been shown to reveal biologically plausible evolutionary phenomena such as horizontal gene transfer between environmentally associated organisms (2, 4, 37, 38). Coordinated loss and divergence of functionally linked genes described here seems to be another such biologically meaningful and potentially important effect. Using sequence similarity as a criterion, it may be difficult to distinguish between “real” gene loss and extreme divergence. Here, we analyzed both phenomena, and we believe that they form a continuum, gene loss being, in many cases, the ultimate case of divergence.
In explaining the observation that S. cerevisiae lacks many genes shared by S. pombe and animals and/or plants, an alternative to gene loss is a relatively recent acquisition of these genes by S. pombe via lateral transfer. To fully assess this possibility, additional fungal genome sequences are needed. However, we believe that this is a less likely explanation of the results than gene loss for the following reasons: (i) there is no direct evidence of gene transfer from plants or animals to yeasts; yeasts are free-living organisms whose lifestyle does not involve close contacts with plants or animals; (ii) relatively recent horizontal transfer would suggest unusually high sequence conservation between the respective proteins from S. pombe and their plant or animal orthologs; no indication of such close connections was found (L.A. and E.V.K., unpublished data); (iii) given the general lack of spatial clustering of functionally linked genes on eukaryotic chromosomes, the horizontal transfer hypothesis will have difficulty with the acquisition of entire functional systems.
To characterize the general impact of lineage-specific gene loss on evolution of eukaryotes, many more genome-scale comparisons are required. Applying the protocol used here to a preliminary comparison of the protein sets of the fly Drosophila melanogaster and the nematode Caenorhabditis elegans suggested significant gene loss in the nematode (E.V.K. and L.A., unpublished data).
Patterns of gene coelimination and codivergence seem to have some predictive value for identifying functionally and physically interacting protein sets. Furthermore, unknown functional complexes, in principle, could be identified within the large, miscellaneous category of lost and diverged genes (Fig. 4). Phylogenetic profiles have been proposed recently as a means to predict functional links between proteins that share similar phyletic distribution and evolutionary pattern (39, 40). The approach described here is conceptually similar but complementary in that it is the absence of a set of genes in an organism that may lead to prediction of new functional connections.
Availability of Complete Results.
A complete, annotated list of S. pombe genes whose orthologs in S. cerevisiae have been lost or highly diverged is available at ftp://ncbi.nlm.nih.gov/pub/koonin/Gene_Losses.
Abbreviations
- E
expectation
- PTGS
posttranscriptional gene silencing
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200346997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200346997
References
- 1.Koonin E V, Mushegian A R, Galperin M Y, Walker D R. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Tatusov R L, Wolf Y I, Walker D R, Koonin E V. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 3.Andersson J O, Andersson S G. Curr Opin Genet Dev. 1999;9:664–671. doi: 10.1016/s0959-437x(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 4.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, et al. Nature (London) 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 5.Kupfer D M, Reece C A, Clifton S W, Roe B A, Prade R A. Fungal Genet Biol. 1997;21:364–372. doi: 10.1006/fgbi.1997.0982. [DOI] [PubMed] [Google Scholar]
- 6.Chervitz S A, Aravind L, Sherlock G, Ball C A, Koonin E V, Dwight S S, Harris M A, Dolinski K, Mohr S, Smith T, et al. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 8.Fitch W M. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 9.Walker D R, Koonin E V. Ismb. 1997;5:333–339. [PubMed] [Google Scholar]
- 10.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz J, Milpetz F, Bork P, Ponting C P. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein J. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 15.Aravind L, Ponting C P. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann K, Bucher P. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 17.Mundt K E, Porte J, Murray J M, Brikos C, Christensen P U, Caspari T, Hagan I M, Millar J B, Simanis V, Hofmann K, et al. Curr Biol. 1999;9:1427–1430. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- 18.Shiyanov P, Nag A, Raychaudhuri P. J Biol Chem. 1999;274:35309–35312. doi: 10.1074/jbc.274.50.35309. [DOI] [PubMed] [Google Scholar]
- 19.Logsdon J M, Jr, Stoltzfus A, Doolittle W F. Curr Biol. 1998;8:R560–R563. doi: 10.1016/s0960-9822(07)00361-2. [DOI] [PubMed] [Google Scholar]
- 20.Schiebel W, Pelissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sanger H L, Wassenegger M. Plant Cell. 1998;10:2087–2101. doi: 10.1105/tpc.10.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cogoni C, Macino G. Nature (London) 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 22.Ketting R F, Haverkamp T H, van Luenen H G, Plasterk R H. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 23.Tuschl T, Zamore P D, Lehmann R, Bartel D P, Sharp P A. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 25.Jacobsen S E, Running M P, Meyerowitz E M. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 26.Zou C, Zhang Z, Wu S, Osterman J C. Gene. 1998;211:187–194. doi: 10.1016/s0378-1119(98)00107-3. [DOI] [PubMed] [Google Scholar]
- 27.Jensen S, Gassama M P, Heidmann T. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- 28.Fink G R. Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- 29.Wassenegger M, Pelissier T. Plant Mol Biol. 1998;37:349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- 30.Jones L, Hamilton A J, Voinnet O, Thomas C L, Maule A J, Baulcombe D C. Plant Cell. 1999;11:2291–2302. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aravind L, Walker D R, Koonin E V. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay H D, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr A M. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horvath M P, Schweiker V L, Bevilacqua J M, Ruggles J A, Schultz S C. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 34.Evans S K, Lundblad V. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 35.Kelly T J, Jallepalli P V, Clyne R K. Curr Biol. 1994;4:238–241. doi: 10.1016/s0960-9822(00)00053-1. [DOI] [PubMed] [Google Scholar]
- 36.Hirt R P, Logsdon J M, Jr, Healy B, Dorey M W, Doolittle W F, Embley T M. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, et al. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 38.Ochman H, Lawrence J G, Groisman E A. Nature (London) 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 39.Pellegrini M, Marcotte E M, Thompson M J, Eisenberg D, Yeates T O. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcotte E M, Pellegrini M, Thompson M J, Yeates T O, Eisenberg D. Nature (London) 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]