Abstract
Phylogenetic analysis of mtDNA sequence data confirms the observation that species diversity in the world's smallest living primate (genus Microcebus) has been greatly underestimated. The description of three species new to science, and the resurrection of two others from synonymy, has been justified on morphological grounds and is supported by evidence of reproductive isolation in sympatry. This taxonomic revision doubles the number of recognized mouse lemur species. The molecular data and phylogenetic analyses presented here verify the revision and add a historical framework for understanding mouse lemur species diversity. Phylogenetic analysis revises established hypotheses of ecogeographic constraint for the maintenance of species boundaries in these endemic Malagasy primates. Mouse lemur clades also show conspicuous patterns of regional endemism, thereby emphasizing the threat of local deforestation to Madagascar's unique biodiversity.
Detailed investigation of species diversity for mouse lemurs (genus Microcebus) and other Malagasy organisms is essential for understanding the evolutionary history of this remarkable island, wherein the vast majority of plants and animals are endemic. It is also essential for evaluating conservation risks for these exceptional organisms. The recognition and definition of new species is not a straightforward task because of the numerous species concepts that have been proposed on both theoretical and operational grounds (1–9). The various criteria that have been proposed for recognizing species can be summarized into three general categories: physical (e.g., morphological distinction), historical (e.g., phylogenetic), and biological (e.g., reproductive isolation). Behavioral, ecological, and geographic barriers can act together or separately to promote the genetic and temporal isolation that mark the speciation process. Thus, it has proven to be virtually impossible to derive a species concept that is at once universally applicable, theoretically consistent, and operationally feasible (10). It is often the case that a species may be recognized by one set of criteria but not another, leaving room for debate among differing theoretical or operational perspectives. Ideally, species recognition can be unambiguously supported by a combination of physical, historical, and biological data.
In this report, we present a phylogenetic analysis of mtDNA sequence data and synthesize the multiple layers of evidence and theoretical perspectives that justify a substantial taxonomic revision of mouse lemurs from western Madagascar (11). We also explore the significance of this taxonomy for established models of speciation, classification, and extinction risk in these primates. Mouse lemurs are the world's smallest living primates, with estimated average annual body mass rarely exceeding 60 g (12–14). Like all other native Malagasy primates (infraorder Lemuriformes), they are endemic to Madagascar. Assessments of their relative abundance in the Malagasy primate fauna (13, 15), ready adaptability to secondary forest and other degraded vegetation (16), and widespread distribution (14) have contributed to the perception that they are among the least threatened of endemic Malagasy mammals (17). In the International Union for the Conservation of Nature/Species Survival Commission action plan for lemur conservation (18) they are given a low priority rating. This assessment of their extinction risk relies, however, on the assumption that individual species are geographically widespread. Thus, it is reasoned that localized environmental destruction is less threatening to mouse lemurs than it is to other more geographically restricted species. The analysis of mouse lemur species diversity presented here indicates that this reasoning is misleading and that several species may be more threatened than previously thought.
The history of mouse lemur taxonomy has contributed to the current view of their biogeographic homogeneity. Although numerous specific names have been proposed for the genus Microcebus, it was considered monotypic by most authorities, containing only the species murinus (19), from the time of its original description in 1795 (20) until the late 1970s. Upon increased research activity and broader geographic sampling of mouse lemur populations, several authorities reached the conclusion that there were actually at least two distinct forms (12–14): murinus, a long-eared gray animal from the western regions of Madagascar, and rufus, a short-eared reddish animal from the east. Martin (13), in particular, made note of the differing habitats and ecological constraints defining the two species, with murinus inhabiting dry deciduous and spiny desert forest and specializing on insectivory, and rufus inhabiting humid rain forest and showing dietary tendencies toward omnivory. Thus, the idea that both ecological and biogeographic mechanisms maintain species separation is an implicit assumption of the two-species taxonomy. The two-species ecogeographic classification remained stable until the present decade, during which time mouse lemurs have been studied intensively (21–27). Among other observations, it has been noted that morphologically distinct rufus forms are known to appear in sympatry with murinus forms at several western sites (28). Schmid and Kappeler (29) realized that two distinct species of Microcebus occur in sympatry in the Kirindy Forest [Kirindy/Centre de Formation Professionnelle Forestiere (CFPF)]: one is the typical murinus of dry forests and the second is a distinctly smaller rufous-colored animal. The authors concluded that the second species fit with the original diagnosis of myoxinus, a name that had been in synonymy for several decades. Subsequently, Zimmermann et al. (30) described a new species, M. ravelobensis, from the northwest. These two studies give empirical weight to the theoretical predictions of accelerated cladogenesis among Malagasy primates wherein geographic microhabitats promote speciation and subsequent interspecific competition (28, 31, 32).
Methods
To examine the question of Microcebus species diversity, we analyze genetic and morphometric patterns for individuals from an extensive array of localities and ecological conditions (Table 1). Specimens from most western localities have been prepared as standard skins for morphological study and accessioned into the collections of the Université d'Antananarivo (Madagascar) and the Field Museum of Natural History. Descriptive morphological analysis of these specimens reveals numerous distinguishing characteristics of size, shape, and coloration that vary consistently among groups (e.g., Table 2), prompting the recognition of three mouse lemur species, M. tavaratra (11), M. sambiranensis (11), and M. berthae (11), and the resurrection of two others from synonymy, M. myoxinus (33) and M. griseorufus (34). Here, a discriminant function analysis of 34 cranial, dental, and external morphological characters [corresponding to character 4 from Table 1 (11) plus characters 1–33 from Table 2 (11)] was executed to test the conclusions of the descriptive study.
Table 1.
Collecting localities with general ecotype description
| Ankarana (13*3′S, 49*3′E)* - dry deciduous forest (180 m) |
| *Campement des Américains: degraded ecotone of open savanna with widely scattered trees |
| Campement des Anglais: natural forest on soil base in valleys between deep canyons |
| Forêt d'Analamahitsy: natural forest on limestone bedrock |
| Manongarivo (14*1′S, 48*15′E) - combination of humid and dry forest elements (360 m) |
| Forêt de Bekolosy: degraded lowland forest |
| RNI d'Ankarafantsika (16*20′S, 46*47′E) - dry deciduous forest (160 m) |
| Ankarokaroka: degraded open understory forest on sandy soil |
| Ampijoroa, Jardin Botanique B: degraded forest on sandy soil |
| Tampolo (17*17′S, 49*25′E) - evergreen littoral forest (5–10 m) |
| RNI de Bemaraha (19*6′S, 44*48′E) - dry deciduous forest (140 m) |
| Aboalimena (19*15′S, 44*27′E) - ecotone between savanna and heavily degraded dry |
| deciduous forest (50 m) |
| Kirindy/CFPF (20*3′S, 44*39′E) - dry deciduous forest (40 m) |
| RS d'Andranomena (20*9′S, 44*33′E) - dry deciduous forest (40 m) |
| near Marofandilia: heavily degraded dry deciduous forest |
| Forêt de Manamby (20*26′S, 44*50′E) - dry deciduous forest (180 m) |
| PN de Ranomafana (21*15′S, 47*27′E) - montane humid forest (1000 m) |
| Forêt de Vohimena (22*40′S, 44*49′E) - transitional dry deciduous/humid forest (730 m) |
| RS de Beza Mahafaly (23*40′S, 44*37′E) - dry thorn scrub or spiny forest (130 m) |
| Ihazoara Valley: degraded spiny forest |
| Mandena (24*58′S, 47*01′E) -evergreen littoral forest (5–20 m) |
Elevation is given in parentheses. RNI, Réserve Naturelle Intégrale; RS, Réserve Spéciale; PN, Parc National.
Table 2.
General morphological description and collecting localities of mouse lemur species from western of Madagascar
| Species | Head & body length, mm | Mass, g | Coloration | Locality |
|---|---|---|---|---|
| M. murinus (11) | 129.4 ± 7.89 | 62.3 ± 4.70 | Grayish-brown back with cinnamon diffused mid-dorsal stripe; head brownish | Andranomena, Vohimena, Kirindy and Manamby |
| M. ravelobensis (9) | 127.3 ± 6.67 | 71.7 ± 15.63 | Mottled rufous back with poorly-marked mid-dorsal stripe; rufous head | Ankarafantsika |
| M. tavaratra (6) | 126.3 ± 9.51 | 61.1 ± 15.65 | Dark brown back with distinct mid-dorsal stripe; distinct rufous head markings | Ankarana |
| M. griseorufus (6) | 123.3 ± 6.38 | 62.6 ± 16.36 | Gray back with cinnamon-brown mid-dorsal stripe; washed rufous markings on head | Beza Mahafaly |
| M. berthae (3) | 92.0 ± 2.65 | 30.6 ± 0.57 | Rufous back with well-defined mid-dorsal line; distinctly brighter rufous on head | Kirindy |
| M. sambiranensis (6) | 116.5 ± 4.14 | 44.1 ± 5.91 | Rufous back with poorly-defined mid-dorsal stripe; head amber | Manongarivo |
| M. myoxinus (15) | 123.7 ± 4.76 | 49.0 ± 6.32 | Rufous-brown back with well-defined reddish-brown mid-dorsal stripe; head with distinct rufous-red markings | Aboalimena and Bemaraha |
Body mass measurements were taken for adults captured throughout the year (and thus all seasons). As body mass can change substantially between wet and dry seasons, linear measurements are more consistent estimators of relative body size. Numbers in parentheses indicate sample size for morphological measurements. Details of data collection are given in Rasoloarison et al. (11).
For those individuals prepared as study skins (referenced in ref. 11), DNA was isolated from organ tissues (liver, spleen, or kidney). All other DNA extractions were taken from ear punches. In all cases, genomic DNA was extracted by using the Qiagen (Chatsworth, CA) DNeasy Tissue Kit (cat. no. 69504), amplified via PCR, cycle sequenced by using a dye terminator sequencing kit (Applied Biosystems), and then analyzed by gel electrophoresis with an Applied Biosystems automated DNA sequencer model 377. In choosing a genetic marker, we initially adopted a null hypothesis of species homogeneity by assuming that traditional taxonomy was valid in describing all western populations as a single species. Therefore, we chose a mtDNA marker likely to show genetic variation at the intraspecific level. A segment of approximately 500 bp of the mitochondrial control region, homologous with the hypervariable region 1 (HV1) region in humans, was sequenced for 118 individual mouse lemurs. High levels of genetic variation for the HV1 marker yields a tree in which internal branches are extremely short and poorly resolved, thus making it impossible to address the question of an east/west biogeographic division among populations. To attempt better resolution of deeper nodes, we subsampled individuals from each of nine well-supported HV1 clades and sequenced them for the more conserved cytochrome oxidase subunit II (684 bp) and cytochrome b (1140 bp) genes. Sequences for the reduced taxon sample are available in GenBank under accession numbers AF285451–AF285491 (HV1), AF285492–AF285527 (COII), and AF285528–AF285568 (cytochrome b). The alignment is available from the communicating author.
Results and Discussion
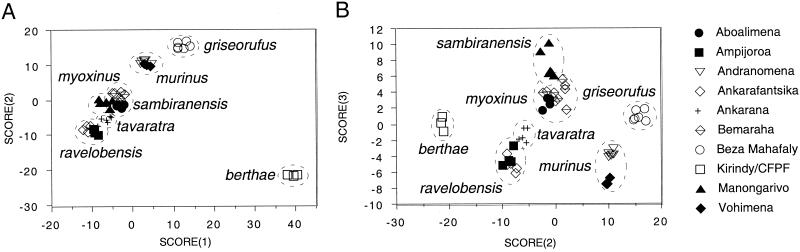
The discriminant function analysis of the morphological data supports the conclusions of the descriptive study (11) by finding that individuals are correctly classified by species designation in 100% of the cases (Fig. 1). Fig. 1A, which compares the first two discriminant functions, distinguishes each of the western mouse lemur species as nonoverlapping clusters. In particular, M. berthae from Kirindy/CFPF is shown to be extremely divergent from the other species. This discrimination is not a simple function of M. berthae's diminutive size. Although M. ravelobensis (the largest species) is loading at the extreme negative end of the scale and M. berthae (the smallest) at the extreme positive end, other species are not assigned according to size, suggesting that function 1 discrimination describes considerable size-independent shape variation among groups. The comparison of the second and third functions (Fig. 1B) further differentiates species on the basis of craniodental shape, indicating that statistical analysis of mouse lemur morphology can be effective for species recognition, although not for reconstructing historical relationships.
Figure 1.
Results from discriminant function analysis of 34 cranial, dental, and external morphometric characters. Body mass was not considered in this analysis. Detailed character descriptions are given in ref. 11. Functions 1 and 2 (A) show conspicuous discrimination of M. berthae from other species. Functions 2 and 3 (B) show discrimination of all species. Combined, the first through third discriminant functions describe 94.5% of the variance in the data set; 55.9%, 31.7%, and 6.9% for the first, second, and third discriminant functions, respectively. Dashed lines are drawn around species clusters for purposes of illustration; they do not convey statistical information.
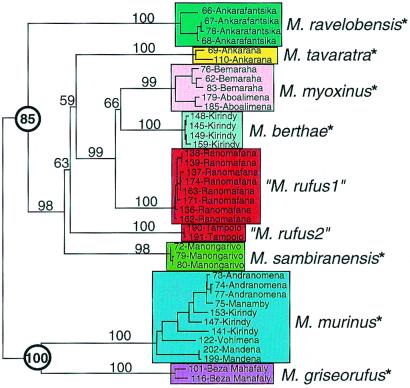
Phylogenetic analysis of suitably variable genetic data can be effective for examining the relationship of taxonomy to phylogeny, assessing historical relationships among putative species, and determining the relationship of that history to current ecological specializations. Parsimony and distance analyses of the HV1 sequences for 118 individuals (not shown) reveal strong support for nine clades. Combined analysis of HV1, COII, and cytochrome b sequences for a reduced sample of 42 individuals reveals nine terminal clades that are identical to those obtained with the HV1 sequences, each resolved with high statistical support (Fig. 2). These clades are also congruent with the various species defined by multivariate analysis of the morphometric data, as would be predicted by the biological species concept (3). Notably, those species that are least defined by the discriminant function analysis (e.g., M. ravelobensis from M. tavaratra; M. sambiranensis from M. myoxinus) are shown to be phylogenetically quite distinct. M. berthae, which is morphometrically most divergent, occupies a nested position in the phylogeny, indicating that its distinguishing morphological characteristics have evolved relatively recently. The results of the combined mtDNA analysis differ from those of the HV1-only analysis in that resolution of deeper branches is far better, showing strong support for two primary clades (bootstrap values highlighted by circles in Fig. 2). Surprisingly, individuals from two eastern localities (Tampolo and Ranomafana), presumed to be representatives of a single species M. rufus, and the sister group to western mouse lemurs, do not form a clade. Rather they form two locality-specific clades that occupy divergent positions within one of the two primary western clades. This result casts doubt on traditional ecogeographic models whereas also suggesting that species diversity may be underestimated for eastern as well as for western mouse lemurs.
Figure 2.
Phylogeny derived from sequence alignment of 2,404 bp of combined mtDNA sequences from the control region homologous with the hypervariable region 1 region in humans, COII and cytochrome b. Clades are color-coded to emphasize species diversity. Individuals are identified by unique laboratory extraction number (Yoder Lab Extraction; YLE) and by locality. Distance tree was generated in paup* 4.0b4a (PPC) (53) by using HKY85 correction model (54) and weighted least squares (power = 2) algorithm. A total of 1,000 replicates of the random addition option were executed without branch swapping. TBR branch swapping then was performed on best tree (hit 399 of 1,000 trials). A single tree of score 0.83 (%SD = 3.27) resulted from the search and is shown with midpoint rooting. Location of midpoint root was confirmed by multiple outgroup rootings. Clade resolution and hierarchy is congruent with strict consensus of 12 trees derived from maximum parsimony analysis in which 100,000 random additions were performed without branch swapping, followed by TBR branch swapping of the 10 best trees. Numbers on branches indicate statistical support from 100 bootstrap replicates with one random addition per replicate. Propithecus, Varecia, and Eulemur were used to root the bootstrap tree. Circled numbers highlight bootstrap support for two primary clades. Asterisks beside species designations indicate species that have been reported to occur in sympatry with another mouse lemur type. See ref. 11 for details.
The analysis of morphological and molecular data presented above shows strong physical and historical evidence for unexpectedly high levels of species diversity among western mouse lemurs. There is also considerable evidence of reproductive isolation to verify these conclusions. Fig. 2 reveals that the Kirindy/CFPF locality harbors representatives from both of the primary mouse lemur clades, M. berthae from one and M. murinus from the other. This phylogenetic separation of haplotypes from sympatrically occurring animals indicates the presence of reproductive barriers to gene flow, an inference that is supported by long-term documentation of noninterbreeding between the two mouse lemur species (29). Sympatric distribution of two or more mouse lemur types also has been reported for the majority of other western localities (affected species are marked with * in Fig. 2), demonstrating that there is strong anecdotal evidence of reproductive barriers among most of the other western species (11, 17, 22, 29, 30, 35–38). Another study has shown that male advertisement calls (a potential reproductive isolating mechanism) can differ significantly, even among genetically and morphologically homogeneous demes within a single M. murinus population (39). In fact, similar isolating mechanisms have been demonstrated to maintain species boundaries in African bushbabies, a closely related and ecologically analogous group of primates (40).
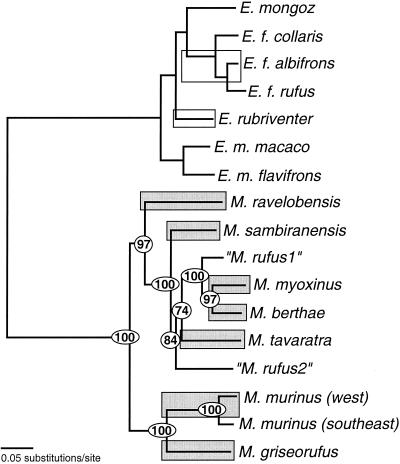
The combined physical, historical, and biological evidence of species diversity in mouse lemurs has important implications for models of Malagasy primate evolution. To investigate questions of when and how the initial mouse lemur radiation occurred, it is desirable to place the inferred speciation events into a temporal framework. By doing so, one can determine the probable geological and environmental context within which those events occurred. The use of genetic data for inferring divergence dates is a common practice, although such studies typically meet two criteria: rates of genetic evolution among organismal lineages are consistent with a molecular clock model [although new methods are also being developed that allow estimation of divergence dates in the absence of a global molecular clock (41–44)] and a reliable fossil record is available for calibrating that clock. Our study is able to meet the demands of the first criterion (Fig. 3), but not the second. Madagascar's fossil record is devoid of vertebrate fossils for the period spanning the late Cretaceous (45, 46) through the late Pleistocene (47), the very period within which lemuriforms are believed to have arrived and diversified (48, 49).
Figure 3.
Illustration of Microcebus mtDNA branch lengths (shaded boxes) compared with those of Eulemur species (open boxes). Two species highlighted for Eulemur (E. fulvus and E. rubriventer) are morphologically distinct and occur in noninterbreeding sympatry. The following individuals (identified by YLE number and by field number) were chosen as representative haplotypes: M. ravelobensis, YLE66 (RMR58); M. sambiranensis, YLE72 (RMR40); “M. rufus1,” YLE138 (SA M102); M. myoxinus, YLE62 (RMR30); M. berthae, YLE148 (JUG 72); M. tavaratra, YLE110 (RMR76); “M. rufus2,” YLE190 (SMG 8774); M. murinus (west), YLE74 (RMR47); M. murinus (east), YLE199 (JUG A-8A1D); M. griseorufus, YLE116 (RMR67). Tree was generated in paup* 4.0b4a (PPC) by using the maximum-likelihood optimality criterion. Settings corresponded to the HKY85 model with rate heterogeneity. -Ln likelihood, 12982.79; estimated ti/tv, 4.60 (kappa, 9.60); estimated gamma shape parameter, 0.26. Circled numbers indicate quartet puzzling values from 1,000 puzzling steps of HKY85 model with sites assumed to evolve at single rate. Likelihood ratio test conducted in puzzle 4.0.1 (55) indicates that a molecular clock cannot be rejected (critical significance level = 14.96%).
There are two potential solutions to the fossil calibration problem. Either outgroups for which there is a detailed fossil record can be included in the study or relative estimates of temporal diversification can be achieved. Fig. 3 illustrates the application of the latter approach. Individual combined mtDNA haplotypes were chosen as representatives from each mouse lemur clade and were analyzed with maximum likelihood, along with homologous sequences for multiple species within the Eulemur radiation. By including the Eulemur taxa, we can compare the depth of mouse lemur diversity to that of a closely related clade for which species diversity is well documented. The resulting tree (Fig. 3), drawn to show proportional branch lengths, illustrates the relative constancy of evolutionary rates within and between the two genus-level clades. The visual impression of uniform rates was confirmed with a likelihood ratio test. This analysis therefore lends even more support to the conclusion of mouse lemur species diversity in that relative branch lengths for two sympatrically occurring Eulemur groups, known to be morphologically and biologically distinct (open boxes), show levels of evolutionary divergence equivalent to those of the seven west coast mouse lemur species (shaded boxes). And, although absolute dates cannot be placed on the nodes of interest because of the paucity of the fossil record, it can still be deduced that the initial Eulemur and Microcebus radiations were approximately contemporaneous, perhaps suggesting a period of climatic variability and associated environmental change in Madagascar.
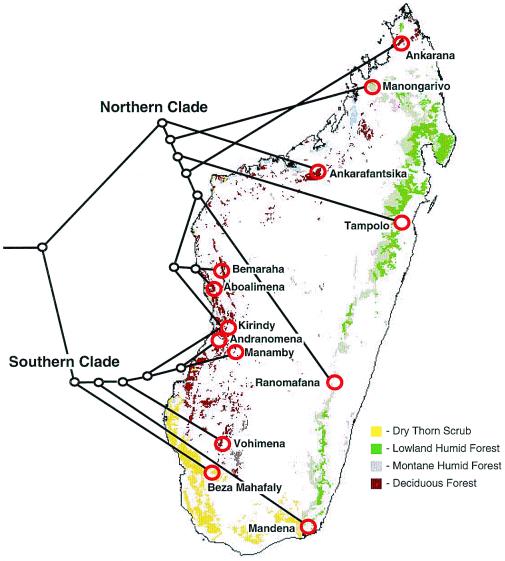
By mapping the hierarchical outline of the mtDNA haplotype phylogeny onto the localities from which the samples were collected (Fig. 4), the hypothesis that there is a fundamental division between eastern wet-adapted and western dry-adapted mouse lemurs can be further tested. Under this ecogeographic model, we would expect to observe haplotypes from eastern localities and western localities segregating into mutually exclusive clades. Such is not the case. Instead, a primary division into northern and southern clades is recovered with both clades comprised of individuals from both wet and dry forest habitats. Thus, there is no phylogenetic basis for classifying mouse lemurs according to their current adaptation to wet versus dry environments. Indeed, ecological plasticity within clades seems to be typical, even at the intraspecific level. For example, the M. murinus clade (Fig. 2) contains individuals native to both dry western (e.g., Andranomena) and distinctly wetter southeastern (e.g., Mandena) environments. In this instance, however, it is likely that a western population colonized wet evergreen forest via transitional littoral forest rather than via the rain forests of the eastern mountain range. The observed distribution of alternating ecotypes within clades is contradictory to recent proposals of ecological niche conservatism during the speciation process (50) as well as for the established east/west biogeographic model. Instead, the phylogeographic pattern recovered by this study offers an alternative hypothesis that there may be or have been a significant biogeographic barrier separating northern and southern lemuriform communities. Our results are therefore compatible with three previous studies of lemuriform distributional patterns that also have postulated a possible biogeographic division between northern and southern communities, with exchange occurring between eastern and western localities (35, 51, 52). The present study is lacking, however, in that M. murinus individuals were not sampled from north of Kirindy/CFPF. Such samples could significantly impact the perceived north/south biogeographic division. Further sampling of eastern localities also will be important for establishing a complete picture of mouse lemur species diversity. Nonetheless, the phylogeographic analysis presented here clearly indicates that mouse lemurs show strong patterns of local endemism, and consequently, some species may be at higher risk for extinction than previously thought. Deforestation at the local level threatens not only populations, but entire species.
Figure 4.
Combined mtDNA haplotype phylogeny (from Fig. 2) superimposed on mouse lemur collecting localities. Figure shows segregation of haplotypes into northern and southern clades with 85% and 100% bootstrap support, respectively. The placement of the root was confirmed by multiple rooting techniques (outgroup and midpoint), a variety of outgroup taxon samples, and three different optimality criteria (maximum parsimony, distance, and maximum likelihood). Note that Kirindy/CFPF contains individuals from both northern and southern clades. Map modified with permission from ref. 56. Ecotype designations are general; for site-specific information, refer to Table 1.
Acknowledgments
We thank J. Coyne, M. Dagosto, J. Flynn, D. Hull, D. Lees, R. D. Martin, L. Olson, and J. G. M. Thewissen for helpful comments on the manuscript and D. Schwab for help in collecting tissue samples. C. Groves provided important information with regard to proper referencing of the new species names. D. Lees generously provided the map shown in Fig. 4. Financial support was provided by Conservation International, Deutsche Forschungsgemeinschaft (Ga 342/3-1,2), the Margot Marsh Biodiversity Foundation, the World Wide Fund for Nature, a Northwestern University Research Grants Committee award, and by National Science Foundation Grant DEB-9985205 (to A.D.Y.).
Abbreviations
- HV1
hypervariable region 1
- CFPF
Centre de Formation Professionnelle Forestiere
- YLE
Yoder Lab Extraction
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF285451–AF285568).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200121897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200121897
References
- 1.Avise J C, Wollenberg K. Proc Natl Acad Sci USA. 1997;94:7748–7755. doi: 10.1073/pnas.94.15.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyne J A, Orr H A. In: Speciation and Its Consequences. Otte D, Endler J A, editors. Sunderland, MA: Sinauer; 1989. pp. 180–207. [Google Scholar]
- 3.Coyne J A, Orr H A. Philos Trans R Soc London B. 1998;353:287–305. doi: 10.1098/rstb.1998.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cracraft J. Curr Ornithol. 1983;1:159–187. [Google Scholar]
- 5.Davis J I, Nixon K C. Syst Biol. 1992;41:421–435. [Google Scholar]
- 6.Donoghue M J. Bryologist. 1985;88:172–181. [Google Scholar]
- 7.Mayr E. Systematics and the Origin of Species from the Viewpoint of a Zoologist. New York: Columbia Univ. Press; 1942. [Google Scholar]
- 8.Mishler B D, Donoghue M J. Syst Zool. 1982;31:491–503. [Google Scholar]
- 9.Templeton A R. In: Molecular Ecology and Evolution: Approaches and Applications. Schierwater B, Streit B, Wagner G P, DeSalle R, editors. Basel: Birkhauser; 1994. pp. 455–477. [Google Scholar]
- 10.Hull D L. In: Species: The Units of Biodiversity. Claridge M F, Dawah H A, Wilson M R, editors. New York: Chapman & Hall; 1997. pp. 357–380. [Google Scholar]
- 11.Rasoloarison R M, Goodman S M, Ganzhorn J U. Int J Primatol. 2000;21:963–1019. [Google Scholar]
- 12.Petter J-J, Albignac R, Rumpler Y. Faune de Madagascar 44: Mammifères Lémuriens (Primates Prosimiens) Vol. 44. Paris: Organisation pour la Recherche Scientifique et Technique Outre Mer and Centre National de la Recherche Scientifique; 1977. pp. 1–513. [Google Scholar]
- 13.Martin R D. Zeitschrift für Tierpychologie. 1972;9,Suppl.:43–89. [Google Scholar]
- 14.Tattersall I. The Primates of Madagascar. New York: Columbia Univ. Press; 1982. [Google Scholar]
- 15.Charles-Dominique P, Hladik C M. La Terre et la Vie. 1971;25:3–66. [Google Scholar]
- 16.Martin R D. In: Comparative Ecology and Behaviour of Primates. Michael R P, Crook J H, editors. London: Academic; 1973. pp. 1–68. [Google Scholar]
- 17.Mittermeier R A, Tattersall I, Konstant W R, Meyers D M, Mast R B. Lemurs of Madagascar. Washington, DC: Conservation International; 1994. [Google Scholar]
- 18.Mittermeier R A, Konstant W R, Nicoll M E, Langrand O. Lemurs of Madagascar: An Action Plan for Their Conservation, 1993–1999. Gland, Switzerland: International Union for the Conservation of Nature/Species Survival Commission Primate Specialist Group; 1992. [Google Scholar]
- 19.Schwarz E. Proc Zool Soc London. 1931;1931:399–428. [Google Scholar]
- 20.Geoffroy Saint-Hilaire E. Bull Soc Philomath Correspondans. 1795;1:89–90. [Google Scholar]
- 21.Atsalis S, Schmid J, Kappeler P M. J Human Evol. 1996;31:61–68. [Google Scholar]
- 22.Ausilio E, Raveloanrinoro G. Lemur News. 1998;3:4–7. [Google Scholar]
- 23.Fietz J. Am J Primatol. 1999;48:127–133. doi: 10.1002/(SICI)1098-2345(1999)48:2<127::AID-AJP4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Ganzhorn J U, Schmid J. Int J Primatol. 1998;19:785–796. [Google Scholar]
- 25.Perret M. J Biol Rhythms. 1997;12:136–145. doi: 10.1177/074873049701200205. [DOI] [PubMed] [Google Scholar]
- 26.Radespiel U, Cepok S, Zietemann V, Zimmermann E. Am J Primatol. 1998;46:77–84. doi: 10.1002/(SICI)1098-2345(1998)46:1<77::AID-AJP6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 27.Rumpler Y, Ganzhorn J U, Tomiuk J, Leipoldt M, Warter S. Folia Primatol. 1998;69:307–311. doi: 10.1159/000021644. [DOI] [PubMed] [Google Scholar]
- 28.Martin R D. In: Creatures of the Dark. Alterman L, Doyle G A, Izard M K, editors. New York: Plenum; 1995. pp. 535–563. [Google Scholar]
- 29.Schmid J, Kappeler P M. Folia Primatol. 1994;63:162–170. doi: 10.1159/000156812. [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann E, Cepok S, Rakotoarison N, Zietemann V, Radespiel U. Folia Primatol. 1998;69:106–114. doi: 10.1159/000021571. [DOI] [PubMed] [Google Scholar]
- 31.Martin R D. Philos Trans R Soc London B. 1972;264:295–352. doi: 10.1098/rstb.1972.0013. [DOI] [PubMed] [Google Scholar]
- 32.Martin R D. Int J Primatol. 2000;21:1021–1049. [Google Scholar]
- 33.Peters W C H. Naturwissenschaftliche Reise nach Mossambique. Berlin: Georg Reimer; 1852. [Google Scholar]
- 34.Kollman M. Bull Museum Natl d'Histoire Naturelle, Paris. 1910;16:301–304. [Google Scholar]
- 35.Thalmann U, Rakotoarison N. Folia Primatol. 1994;63:156–161. doi: 10.1159/000156811. [DOI] [PubMed] [Google Scholar]
- 36.Randrianambinina B. Contribution à l'étude biologique de Microcebus ravelobensis dans la région d'Ampijoroa/Ankarafantsika. Antananarivo: Université d'Antananarivo; 1997. [Google Scholar]
- 37.Nicoll M E, Landgrand O. Madagascar: Revue de la conservation et des aires protégées. Gland: World Wide Fund for Nature; 1989. [Google Scholar]
- 38.Rakotoarison N, Mutschler T, Thalmann U. Oryx. 1993;27:35–40. [Google Scholar]
- 39.Hafen T, Neveu H, Rumpler Y, Wilden I, Zimmermann E. Folia Primatologica. 1998;69, Suppl. 1:342–356. doi: 10.1159/000052723. [DOI] [PubMed] [Google Scholar]
- 40.Bearder S K, Honess P E, Ambrose L. In: Creatures of the Dark. Alterman L, Izard M K, Doyle G A, editors. New York: Plenum; 1995. pp. 331–352. [Google Scholar]
- 41.Huelsenbeck J P, Larget B, Swofford D. Genetics. 2000;154:1879–1892. doi: 10.1093/genetics/154.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorne J L, Kishino H, Painter I S. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z. Comput Appl Biosci. 1997;15:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 44.Sanderson M J. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- 45.Krause D W, Prasad G V R, von Koenigswald W, Sahni A, Grine F E. Nature (London) 1997;390:504–507. [Google Scholar]
- 46.Krause D W, Hartman J H, Wells N A. In: Natural Change and Human Impact in Madagascar. Goodman S M, Patterson B D, editors. Washington, DC: Smithsonian Institution; 1997. pp. 3–43. [Google Scholar]
- 47.Burney D A. In: Natural Change and Human Impact in Madagascar. Goodman S M, Patterson B D, editors. Washington, DC: Smithsonian Institution; 1997. pp. 75–89. [Google Scholar]
- 48.Goodman M, Porter C A, Czelusniak J, Page S L, Schneider H, Shoshani J, Gunnell G, Groves C P. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 49.Yoder A D, Cartmill M, Ruvolo M, Smith K, Vilgalys R. Proc Natl Acad Sci USA. 1996;93:5122–5126. doi: 10.1073/pnas.93.10.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson A T, Sober-n J, Sánchez-Cordero V. Science. 1999;285:1265–1267. doi: 10.1126/science.285.5431.1265. [DOI] [PubMed] [Google Scholar]
- 51.Ganzhorn J U. Folia Primatol. 1998;69:332–341. doi: 10.1159/000021644. [DOI] [PubMed] [Google Scholar]
- 52.Pollock J I. Primate Conserv. 1986;7:82–86. [Google Scholar]
- 53.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 54.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 55.Strimmer K, von Haesler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 56.Lees D C, Kremen C, Andriamampianina L. Biol J Linnean Soc. 1999;67:529–584. [Google Scholar]