Abstract
The four species of “river dolphins” are associated with six separate great river systems on three subcontinents and have been grouped for more than a century into a single taxon based on their similar appearance. However, several morphologists recently questioned the monophyly of that group. By using phylogenetic analyses of nucleotide sequences from three mitochondrial and two nuclear genes, we demonstrate with statistical significance that extant river dolphins are not monophyletic and suggest that they are relict species whose adaptation to riverine habitats incidentally insured their survival against major environmental changes in the marine ecosystem or the emergence of Delphinidae.
Cetaceans are ecologically diverse, because their habitats range from coastal to oceanic and from tropical to polar waters. Whereas most of the 83 recognized extant species (1) are exclusively marine, several species live sporadically or exclusively in freshwater. Some of them (e.g., Beluga, Tucuxi, Irrawaddy dolphin, and finless porpoise) are well characterized phylogenetically, i.e., they belong to the superfamily Delphinoidea (white whales, true dolphins, and porpoises) (2, 3). On the other hand, the taxonomic relationships of the so-called river dolphins have been in a state of confusion for more than a century. These include three exclusively riverine species: (i) the blind river dolphin, or “susu” (Platanista gangetica) living in the Indus, Ganges, and Brahmaputra river systems on the Indian subcontinent; (ii) the Yangtze river dolphin, or “baiji” (Lipotes vexillifer), which lives in the lower and middle reaches of the Yangtze river in China, and (iii) the Amazon river dolphin, or “boto” (Inia geoffrensis), which is largely distributed in northern South America in the Orinoco and Amazon River systems, and the upper Rio Madeira drainage. The fourth species classified as a river dolphin is the La Plata dolphin, or “franciscana” (Pontoporia blainvillei). It is found not only in estuaries but also in coastal waters of eastern South America from 19°S (Brazil) to 42°S (Argentina).
Most populations of the four species are decreasing because of multiple threats such as direct or incidental catch, hydroelectric development, collisions with boats, deforestation, and pollution from agriculture, industry, and mining. The susu and baiji are the most endangered, with the latter probably represented by less than 100 individuals so that there is very little hope for its short-term survival (4).
All river dolphins show a peculiar morphology with a characteristic long and narrow rostrum, a low triangular dorsal fin, broad and visibly fingered flippers, and a flexible neck. Their eyes have also been reduced to various degrees (ref. 5; the susu even lacks eye lenses and is virtually blind) whereas their echolocation abilities could be more refined than in other cetaceans. In addition, shared skull characters led most authors to classify them into a monophyletic group, either in the family Platanistidae or in the superfamily Platanistoidea (sensu lato; refs. 6–11). However, these diagnosing characters could be ancestral (1, 12), hence phylogenetically uninformative, or prone to convergence (because adaptive to living in turbid waters). Whereas the monophyly of the group was usually not questioned, many morphological analyses emphasized the substantial divergence among the four species (13, 14), and this eventually led to the classification of the four genera in four monotypic families (15, 16). Morphological idiosyncrasies as well as the separation of the four species on three subcontinents have created taxonomic disagreement among morphologists with continuing debate regarding the reality of a river dolphin clade and the position(s) of river dolphins within the phylogeny of whales. Gray (17) first challenged the monophyly of river dolphins and was followed by other authors who included the fransiscana within delphinids (18, 19). Only recently, morphologists revisited Gray's hypothesis. Heyning (2) recognized a clade including the boto, fransiscana, and baiji. On the other hand, de Muizon (20, 21), as well as Messenger and McGuire (22), grouped the boto and fransiscana in a clade closely related to the Delphinoidea, and suggested that the baiji is the sister group of this [boto + fransiscana + Delphinoidea] clade.
Unfortunately, current paleontological data cannot resolve the issue because the fossil record of river dolphins is rather scarce, geographically isolated, and dates back to the Miocene and lower Pliocene. Furthermore, different authors disagree both on the ecological (marine or riverine) and taxonomic status of potentially key fossil specimens (11, 20, 23, 24), making alternative hypotheses on how and when river dolphins invaded riverine habitats controversial (see refs. 25 and 26 for conflicting hypotheses regarding the boto). On the other hand, it is well established on the basis of paleontological data that various extinct groups, several of which are closely related to the Platanistidae (i.e., the lineage to which the susu belongs), underwent a considerable adaptive radiation (Fig. 2), were globally distributed, and included numerous species representing different ecomorphs (21, 27–29). Molecular analyses so far did not fully resolve the issue either because they lacked one or several extant river dolphin species (30, 31).
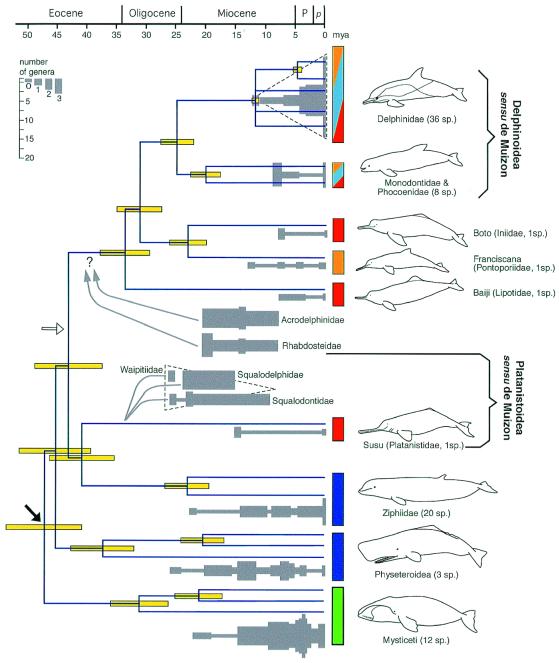
Figure 2.
ML tree topology (−lnL = 19019.05697) combined with calculated dates of divergences as well as estimates of generic diversity of relevant fossil taxa (gray-shaded boxes). Generic diversity estimates are from ref. 32. Only genera considered stable enough and based on diagnostic material are considered. Numbers of extant species are given between parentheses for each group. Vertical colored boxes indicate habitat: green, specialization in filter feeding (baleen whales); dark blue, specialization for deep feeding on squids (beaked and sperm whales); orange, coastal waters; light blue, oceanic waters; and red, fluvial environment (whereas the boto, baiji, and susu are exclusively riverine, some delphinoid species consist of distinct coastal and fluvial populations). Each horizontal yellow bar indicates twice the value of the standard deviation for the age of the corresponding node. Hence, standard deviations are not referring to the topology of the tree. Dotted triangles indicate the radiation of Delphinidae and concurrent gradual extinction of Waipitiidae, Squalodelphidae, and Squalodontidae. The black arrow at the base of the tree indicates the position of the root when all nu and mt data are used in a simultaneous ML analysis and by using hippopotamus as an outgroup. This result is, however, unstable because several gene fragments support an alternative root, indicated by the white arrow. Whichever of these two rooting hypotheses is correct is irrelevant to the issues discussed and conclusions reached in the present paper. P, Pliocene and p, Pleistocene.
By using mitochondrial (mt) and nuclear (nu) DNA sequences from all extant river dolphin species and representatives of all other major cetacean groups, we performed phylogenetic analyses, calculated the age of nodes on the inferred trees, and combined these results with abundance estimates on relevant fossil cetacean taxa (32). Because the assumption of similar rates of evolution among different lineages is unrealistic, we extracted dating information from our data set with a recently developed approach (33), which is explicitly independent of any molecular clock.
Materials and Methods
We based our phylogenetic analyses and dating of evolutionary events on nucleotide sequences from three mt and two nu genes from 19 cetacean species and one outgroup. The target mtDNA fragments were (i) a 382–404-bp segment of the 12S rRNA gene (primers for PCR and sequencing from ref. 31); (ii) a 505–548-bp segment of the 16S rRNA gene (primers from ref. 31); and (iii) the full cytochrome b gene (and flanking regions) (primers CB-out1, 5′-AATGAYATGAAAARYCATCGTTG-3′; CB-out2, 5′-TCTTCCTTGAGTCTTAGGGAG-3′; CB-in1, 5′-TTRTTRGATCCTGTTTCRTG-3′; and CB-in2, 5′-TGAGGACAAATATCATTYTGAG-3′). The nuDNA fragments were (i) a 1,079–1,106-bp segment of the gene encoding the interphotoreceptor retinoid-binding protein (primers 1F, 5′-GCCTGGTCATCTCCTATGAGCC-3′; 1R, 5′-CAGACTGGCCTATCCTCAGCTTC-3′; 2F, 5′-GTCCTCACCAGTGGTCGCA-3′; 2R, 5′-GGCCGGTGTTGAGCTTAGTG-3′; 3F, 5′-GCTGCCTTGTGTGGGGACAC-3′; 3R, 5′-TGTCCTGCAGGGGCTCCCACA-3′; and others from ref. 34), and (ii) a 1,021–1,140-bp segment of the lactalbumin gene (primers from ref. 35). To help rooting the phylogenetic tree, we additionally sequenced fragments of the transferrin (primers TRSFEX6F, 5′-TGAAGGATGGTGCTGGGGATGTGGCCTT-3′; and TRSFEX8R, 5′-ACCCGTGGGCAGAGTCCTTAAACAGCA-3′) and the von Willebrand factor (primers from ref. 36) genes (1,489–1,539 bp and 931–1,237 bp, respectively) in Hippopotamus amphibius and several ingroup taxa. The DNA fragments were PCR amplified and sequenced (dRhodamine Cycle Sequencing; Applied Biosystems) on both strands; sequencing of complementary strands was performed on different PCR products. The accession numbers of the published and new sequences are all indicated in Table 1 together with the species to which they correspond.
Table 1.
Binomial and common species names, gene fragments, and corresponding GenBank accession nos. of sequences included in the analyses
Data from transferrin and von Willebrand factor genes were used only in analyses by which we attempted to root the cetacean tree; sequences not available for some of the ingroup taxa were then treated as missing data.
Sequences new to this study.
To avoid artifactual results because of ambiguity in alignment, we used the program soap (A. Löytynoja and M.C.M., http://dbm.ulb.ac.be/ueg) to produce one alignment for each of 25 different sets of alignment parameters (weighted matrix, gap penalties from 11 to 19 by steps of 2, and extension penalties from 3 to 11 by steps of 2). Positions at which alignments differed were excluded (37). This sensitivity analysis yielded a data matrix of 4,437 unambiguously aligned characters. All alignments are available on request from M.C.M. Gaps resulting from the alignment were treated as missing data.
Partition homogeneity tests under maximum parsimony (MP) (38) on interphotoreceptor retinoid-binding protein vs. lactalbumin and nu vs. mt indicated not significant incongruence among these data sets as soon as the clearly saturated mt transition substitutions were excluded. Three data sets were analyzed: (i) mtDNA; (ii) nuDNA; and (iii) “total DNA” evidence, i.e., concatenation of the two former data sets. paup* (39) was used for all phylogenetic analyses. MP analyses were first performed with all characters weighted equally. Stability of the cladograms was then tested with the Goloboff fit criterion (40) (with k = 0, 2, 4, and 8), which allows individual downweighting of noisy characters. We also used the maximum likelihood (ML) method of phylogeny inference with the following settings (paup*): empirical nucleotide frequencies, transition/transversion ratio (Ti/Tv), and proportion of invariable sites estimated via ML, Hasegawa-Kishino-Yano model (41) with rate heterogeneity, rates for variable sites assumed to follow a γ distribution with shape parameter estimated by ML, and tree bisection-reconnection branch-swapping. For neighbor-joining (NJ) analyses, LogDet distances (42) were calculated, after removing different proportions of invariable sites, to correct for possible differences in base composition among lineages. The stability of individual clades was estimated by computing bootstrap values (43) for NJ, MP, and ML trees. Given the high computation burden of the latter tree inference method, it was practical to perform a ML bootstrap analysis (400 replicates) only by constraining ML parameters values (Pinv, Ti/Tv ratio, and γ shape) to those obtained in the ML search on the original (nonresampled) data set. Specific alternative hypotheses were compared statistically under ML and MP by means of Kishino-Hasegawa tests (44). Furthermore, we performed T-PTP tests (45) to examine whether the support of the observed data for a given branch is significantly different from the support achievable through randomization of the data. Using a priori T-PTP is valid because we used the test only to evaluate hypotheses of monophyly/nonmonophyly formulated before any of our phylogenetic analyses.
We extracted dating information (and standard deviations on these estimates) from our data set with the programs estbranches and divtime5, implementing a recently developed Bayesian approach (33), which allows variation of molecular rates across the phylogenetic tree. Calibration was performed by constraining the origin of delphinids to have happened between 11 and 13 million years ago (24, 32, 46). We computed both the prior and posterior distributions to verify that the data provide significant information to the estimation of divergence time. We also computed the posterior twice (i.e., by using two different random seeds) to check for convergence of the results.
Results and Discussion
Our MP, ML, and NJ analyses indicate that river dolphins are polyphyletic. Constraining the monophyly of that group requires a significant reduction in likelihood (P < 0.001, P < 0.0002, and P < 0.0001, for the nu, mt, and combined data sets, respectively) and a significant increase in tree length under unweighted MP (P < 0.0026). Analyses of the full data set under NJ, MP, and ML yielded each a tree (Figs. 1 and 2) compatible with recent morphological findings (2, 20–22), suggesting that river dolphins are neither monophyletic nor included within Delphinoidea. More specifically, our analyses indicate that (i) the two South American river dolphins (boto and fransiscana) form the sister group of the Delphinoidea; (ii) the baiji is the sister species of this [Delphinoidea + boto + fransiscana] clade, providing additional evidence for the results of de Muizon (20, 21) and Messenger and McGuire (22). This is consistent with the fact that fossils related to the iniid and pontoporiid families were found in the same layers of the La Plata region (11). However, we could not reject with statistical significance the possibility of a monophyletic [boto + fransiscana + baiji] (2).
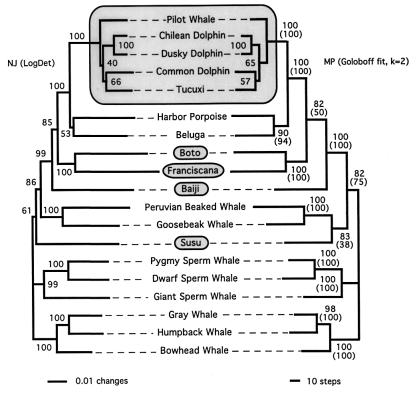
Figure 1.
Unrooted molecular phylogeny of cetaceans based on nuDNA and mtDNA sequences. The left and right sides show the NJ (under LogDet distances) and MP (under Goloboff fit criterion with k = 2) trees, respectively. Bootstrap values are indicated at the nodes (1,000 and 400 replicates for NJ and MP, respectively). Values between parentheses correspond to bootstrap values (400 replicates) for ML analyses (see Fig. 2 for tree topology). Topology of the MP tree is stable to Goloboff weighting (with k = 0–8). Unweighted MP analysis yielded a tree in which the positions of the sperm whale and [beaked whales + susu] clades are exchanged; these two alternative topologies have tree lengths differing by two evolutionary events and are not significantly different under the Kishino-Hasegawa test. The large and small shaded boxes indicate the radiation of delphinids and the four river dolphin species, respectively. Note that the exact placement of the susu differs between the NJ and MP/ML trees.
In agreement with many authors, we find the susu to be more basal than the other river dolphin species. This is compatible with the description of several ancestral features in this species (5) and with the suggestion that Miocene-extinct Squalodontidae, Squalodelphidae, and Waipatiidae have close phylogenetic affinities with Platanistidae (21, 27). We will refer to this assemblage of four families as Platanistoidea, sensu de Muizon (Fig. 2). We could not resolve with high significance the exact phylogenetic position of the susu for probably two reasons. First, the susu, sperm, beaked, and baleen whale lineages seem to have been produced through a very rapid succession of splitting events in the Eocene as indicated by our estimates of divergence time (Fig. 2). This might explain that the placement of the susu lineage with beaked whales or at the base of a [beaked whales + baiji + boto + fransiscana + dephinoids] clade (compare Figs. 1 and 2) could not be supported by more than three among the nearly 5,000 analyzed characters. Second, there is conflicting signal between the nuDNA and mtDNA data. This latter problem is particularly prominent when we attempt to root the cetacean tree (Fig. 2), and it could be responsible for the major controversy regarding the possible nonmonophyly of toothed whales (cf. refs. 22 and 47). Whether this conflict is caused by differential lineage sorting or by misleading signal from one or several data set(s) remains to be investigated and is beyond the scope of the present article.
Combination of our molecular analyses on tree inference and assessment of splitting dates with paleontological estimates of generic diversity suggests that extant river dolphins are relict species (Fig. 2). Indeed, the fossil record indicates that the susu lineage belongs to the assemblage of Platanistoidea (sensu de Muizon), which experienced a considerable and global radiation (refs. 21, 27, and 48; Fig. 2). Platanistoidea as well as Acrodelphinidae and Rhabdosteidae (Fig. 2) are characterized by a remarkable variety of long-snouted dolphins including highly specialized and aberrant forms (49). As the two latter groups are probably not specifically related to any river dolphin lineage, the long-beaked morphotype is either ancestral or convergent.
In the middle Miocene, platanistoids (sensu de Muizon) started to decline whereas the number of delphinoid species increased dramatically (27–29, 48, 50), raising the possibility that the latter ecologically displaced the former (28, 48). However, such explanation involving competition among groups has been questioned for many replacement events, which, alternatively, could either correspond to an emerging pattern caused by random processes of extinctions and radiations, or be the result of major environmental changes (refs. 51 and 52 and refs. therein). Similarly, the replacement of platanistoid by delphinid dolphins might have occurred without direct competition between the two groups. Worldwide changes in the environmental conditions might have caused the extinction of the Miocene platanistoid dolphins, but also the concurrent radiation of modern oceanic dolphins into an increasing variety of habitats (refs. 28 and 48 for details). Furthermore, competitive takeovers and extinction/opportunistic replacement events could be intermingled because competition may increase the likelihood of extinction of a lineage already impaired by external physical changes.
With 36 recognized species, modern delphinids represent the most successful and ecologically diverse family of marine mammals (1). All other extant cetacean groups are highly specialized (Fig. 2): beaked and sperm whales occupy rather narrow food niches because they are adapted for deep-feeding on squids, whereas baleen whales developed remarkable filter-feeding abilities which allowed them to exploit food resources not available to other cetaceans, hence, to experience a radiation in the Miocene. A similar reasoning can be used for river dolphins. For example, the susu is the only living species of a once diverse marine taxon (see above). We suggest its ancestor escaped extinction specifically because it was already adapted to the riverine environment, and therefore did not experience competitive interactions with delphinid dolphins and/or was less impacted by changes in the marine environment. As shown in Fig. 2, our dating estimates indicate that all river dolphin lineages diverged well before the radiation of delphinids. It is therefore possible that not only the susu but all living river dolphins are relict species of otherwise successful Oligocene/Miocene groups, which experienced a similar decrease in species number than the platanistoids. Unfortunately, although there was probably a much higher diversity of extinct freshwater dolphins than currently known, fossils are less likely to be found in freshwater deposits than in marine ones and most riverine specimens described so far are too fragmentary to be identified even to family level (J. G. M. Thewissen, personal communication).
Because the franciscana is mostly marine and diverged from the boto well before the radiation of delphinids, it is ambiguous whether the common ancestor of the two South American river dolphins was marine or riverine. If it was indeed riverine, then one must assume a recent ecological reversal, i.e., reinvasion of the marine coastal habitat. On the other hand, the ancestors of the franciscana might have escaped extinction, even though it had not invaded the riverine habitat per se, because it was ecologically specialized. The very small size of the extant species might then reflect coastal adaptations, which would explain its survival in the marine habitat. Furthermore, because we could not reject with statistical significance the possibility of a monophyletic [boto + fransiscana + baiji], a yet older invasion of the riverine habitat could be suggested. However, even under the hypothesis (2) of a monophyletic origin of the extant river dolphin species from South America and China, and despite the paucity of paleontological data on these lineages, an invasion of the riverine environment by their most recent common ancestor would be very difficult to reconcile with plate tectonic data and distribution of the extant species. Hence, all phylogenetic hypotheses compatible with our molecular analyses suggest a minimum of three independent invasions of the freshwater habitat.
In conclusion, we think that the so-called river dolphins is a wastebasket nonmonophyletic (most probably polyphyletic) taxon consisting of relict dolphin lineages from the Eocene, Oligocene, and early Miocene, that developed ecological extreme specialization (including for at least three of the four lineages, adaptation to the fluvial habitat) which incidentally insured their survival against either major changes of physical parameters in the marine environment or the emergence and radiation of better adapted and more recent marine small cetaceans.
Acknowledgments
We thank J. G. M. Thewissen who provided us with unpublished abundance data (32) on the relevant fossil records and insightful comments on Fig. 2; Jeffrey L. Thorne for providing the latest versions of the programs implementing his Bayesian approach to divergence time estimation as well as with advice on its proper use; and Martine Bérubé for providing the two sperm whale lactalbumin sequences. Critical but constructive comments from two anonymous reviewers greatly improved the manuscript. I.C. is supported by the Deutscher Akademischer Austauschdienst Doktorandenstipendium im Rahmen des gemeinsamen HSP III von Bund und Ländern and the Rotary Club Brussels-Erasmus. I.C., S.V., D.V.B., and M.C.M. are supported by the Belgian National Fund for Scientific Research, the Free University of Brussels (ULB), the Van Buuren Fund, the Communauté Française de Belgique (ARC 98/03–223), and the Defay Fund.
Abbreviations
- MP
maximum parsimony
- ML
maximum likelihood
- NJ
neighbor joining
- mt
mitochondrial
- nu
nuclear
Footnotes
References
- 1.Rice D W. Marine Mammals of the World: Systematics and Distribution. Lawrence, KS: Allen; 1998. [Google Scholar]
- 2.Heyning J E. Contrib Sci Nat Hist Mus Los Angeles. 1989;405:1–64. [Google Scholar]
- 3.LeDuc R G, Perrin W F, Dizon A E. Mar Mamm Sci. 1999;15:619–648. [Google Scholar]
- 4.Chen P, Liu R, Wang D, Zhang X. Biology, Rearing and Conservation of Baiji. Beijing: Science Press; 1997. [Google Scholar]
- 5.Pilleri G. Invest Cetacea. 1974;4:44–70. [Google Scholar]
- 6.Flower W F. Trans Zool Soc London. 1869;6:87–116. [Google Scholar]
- 7.Winge H. Smithsonian Misc Collect. 1921;72:1–81. [Google Scholar]
- 8.Slijper E. Capita Zool. 1936;7:1–590. [Google Scholar]
- 9.Simpson G G. Bull Am Museum Nat Hist. 1945;85:1–350. [Google Scholar]
- 10.Gaskin D E. Oceanogr Mar Biol Annu Rev. 1976;14:247–346. [Google Scholar]
- 11.Cozzuol M A. Invest Cetacea. 1985;7:39–53. [Google Scholar]
- 12.Messenger S L. Proc San Diego Soc Nat Hist. 1994;29:125–133. [Google Scholar]
- 13.Kasuya T. Sci Rep Whales Res Inst Tokyo. 1973;25:1–103. [Google Scholar]
- 14.Zhou K, Qian W, Li Y. Acta Zool Sinica. 1979;25:58–74. [Google Scholar]
- 15.Pilleri G, Gihr M. Invest Cetacea. 1980;11:33–36. [Google Scholar]
- 16.Zhou K. Sci Rep Whales Res Inst. 1982;34:93–108. [Google Scholar]
- 17.Gray J E. Proc Zool Soc (London) 1863;31:197–202. [Google Scholar]
- 18.Kellogg R. Q Rev Biol. 1928;3:29–76. , 174–208. [Google Scholar]
- 19.Miller G S., Jr Smithsonian Misc Collect. 1923;76:1–71. [Google Scholar]
- 20.De Muizon C. Comptes Rendus Hebdomadaires des Seances de l'Academie des Sciences, Paris. 1985;5:359–362. [Google Scholar]
- 21.De Muizon C. Proc San Diego Soc Nat Hist. 1994;29:135–146. [Google Scholar]
- 22.Messenger S L, McGuire J A. Syst Biol. 1998;47:90–124. doi: 10.1080/106351598261058. [DOI] [PubMed] [Google Scholar]
- 23.Barnes L G. Contri Sci Natl Hist Museum LA. 1985;363:1–34. [Google Scholar]
- 24.Barnes L G. In: Mammals: Notes for a Short Course, Studies in Geology. Broadhead T W, editor. Vol. 8. Knoxville: University of Tennessee; 1984. pp. 139–158. [Google Scholar]
- 25.Grabert H. Z Saeugetierkunde. 1983;49:334–341. [Google Scholar]
- 26.Lovejoy N R, Bermingham E, Martin A P. Nature (London) 1998;396:421–422. [Google Scholar]
- 27.Fordyce R E. Proc San Diego Soc Nat Hist. 1994;29:147–176. [Google Scholar]
- 28.Whitmore F C. Proc San Diego Soc Nat Hist. 1994;29:223–227. [Google Scholar]
- 29.Gottfried M D, Bohaska D J, Whitmore F C. Proc San Diego Soc Nat Hist. 1994;29:229–238. [Google Scholar]
- 30.Arnason U, Gullberg A. Mol Biol Evol. 1996;13:407–417. doi: 10.1093/oxfordjournals.molbev.a025599. [DOI] [PubMed] [Google Scholar]
- 31.Milinkovitch M C, Meyer A, Powell J R. Mol Biol Evol. 1994;11:939–948. doi: 10.1093/oxfordjournals.molbev.a040164. [DOI] [PubMed] [Google Scholar]
- 32.Thewissen J G M. In: Encyclopedia of Marine Mammals. Perrin W F, Wuersig B, Thewissen J G M, editors. London: Academic; 2001. , in press. [Google Scholar]
- 33.Thorne J L, Kishino H, Painter I S. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 34.Stanhope M J, Czelusniak J, Si J-S, Nickerson J, Goodman M. Mol Phylogenet Evol. 1992;1:148–160. doi: 10.1016/1055-7903(92)90026-d. [DOI] [PubMed] [Google Scholar]
- 35.Milinkovitch M C, Bérubé M, Palsbøll P J. In: The Emergence of Whales: Evolutionary Patterns in the Origin of Cetacea. Thewissen J G M, editor. New York: Plenum; 1998. pp. 113–131. [Google Scholar]
- 36.Porter C A, Goodman M, Stanhope M J. Mol Phylogenet Evol. 1996;5:89–101. doi: 10.1006/mpev.1996.0008. [DOI] [PubMed] [Google Scholar]
- 37.Gatesy J, DeSalle R, Wheeler W. Mol Phylogenet Evol. 1993;2:152–157. doi: 10.1006/mpev.1993.1015. [DOI] [PubMed] [Google Scholar]
- 38.Farris J S, Källersjö M, Kluge A G, Bult C. Cladistics. 1994;10:315–319. [Google Scholar]
- 39.Swofford, D. L. (2000) paup*: Phylogenetic Analysis Using Parsimony (and Other Methods (Sinauer, Sunderland, MA), Version 4.0b4a (in progress).
- 40.Goloboff P A. Cladistics. 1993;9:83–91. doi: 10.1111/j.1096-0031.1993.tb00209.x. [DOI] [PubMed] [Google Scholar]
- 41.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 42.Lockhart P J, Steel M A, Hendy M D, Penny D. Mol Biol Evol. 1994;12:503–513. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- 43.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 44.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 45.Faith D P. Syst Biol. 1991;40:366–375. [Google Scholar]
- 46.McLeod S A, Whitmore F C, Barnes L G. In: The Bowhead Whale. Burns J J, Montague J J, Cowles C J, editors. Lawrence, KS: Allen; 1993. pp. 45–70. [Google Scholar]
- 47.Milinkovitch M C. Trends Ecol Evol. 1995;10:328–334. doi: 10.1016/s0169-5347(00)89120-x. [DOI] [PubMed] [Google Scholar]
- 48.Fordyce R E, Barnes L G. Annu Rev Earth Planet Sci. 1994;22:419–455. [Google Scholar]
- 49.Pilleri G. Mem Sci Geol. 1985;36:1–250. [Google Scholar]
- 50.Barnes L G, Doming D P, Ray C E. Mar Mamm Sci. 1985;1:15–53. [Google Scholar]
- 51.Gould S J, Calloway C B. Paleobiology. 1980;6:383–407. [Google Scholar]
- 52.Benton M J. Nature (London) 1983;302:16–17. [Google Scholar]