Abstract
Exons of three genes were identified within the 85-kilobase tandem triplication unit of the slow Wallerian degeneration mutant mouse, C57BL/WldS. Ubiquitin fusion degradation protein 2 (Ufd2) and a previously undescribed gene, D4Cole1e, span the proximal and distal boundaries of the repeat unit, respectively. They have the same chromosomal orientation and form a chimeric gene when brought together at the boundaries between adjacent repeat units in WldS. The chimeric mRNA is abundantly expressed in the nervous system and encodes an in-frame fusion protein consisting of the N-terminal 70 amino acids of Ufd2, the C-terminal 302 amino acids of D4Cole1e, and an aspartic acid formed at the junction. Antisera raised against synthetic peptides detect the expected 43-kDa protein specifically in WldS brain. This expression pattern, together with the previously established role of ubiquitination in axon degeneration, makes the chimeric gene a promising candidate for Wld. The third gene altered by the triplication, Rbp7, is a novel member of the cellular retinoid-binding protein family and is highly expressed in white adipose tissue and mammary gland. The whole gene lies within the repeat unit leading to overexpression of the normal transcript in WldS mice. However, it is undetectable on Northern blots of WldS brain and seems unlikely to be the Wld gene. These data reveal both a candidate gene for Wld and the potential of the WldS mutant for studies of ubiquitin and retinoid metabolism.
The slow Wallerian degeneration mouse, C57BL/WldS, carries a dominant mutation that delays Wallerian degeneration in the distal stump of an injured axon (1). Wallerian degeneration is a model for the mechanism of axon death in many diseases of the central and peripheral nervous system such as multiple sclerosis, amyotrophic lateral sclerosis, and peripheral neuropathies (2–4), but the mechanism is little understood. It has been suggested that Wallerian degeneration involves an apoptosis-like mechanism (5), although it appears not to involve activation of caspase 3 (6). Other suggestions include activation of calpain (7) and loss of trophic support from the cell body, but at present there are no clear indications as to the key initiating or controlling events. The existence of a mouse mutant indicates that the process is genetically regulated and should make possible the identification of a controlling gene. The WldS mutation has been mapped to distal mouse chromosome 4 (8), and a tandem triplication of an 85-kb genomic region has been identified within a genetic candidate interval (9). The mutation is unique to WldS mice and therefore a strong candidate for the causative change.
The triplication may affect the Wld gene in a number of different ways, depending largely on the relative location of the gene and the triplication. A gene whose full coding and regulatory sequences are located within the repeat unit is likely to overexpress the normal mRNA and protein, as occurs with PMP-22 in Charcot–Marie–Tooth disease type 1a (10). A gene that crosses a boundary of the repeat unit would be disrupted and could encode a truncated protein or a protein fused to the product of another gene. An example would be the fusion between PML and RARα in the chromosomal translocation t(15;17), which causes acute promyelocytic leukemia (11). In each of these situations, a complete exon map of the repeat unit should reveal the causative gene. We cannot rule out the possibility that the causative gene lies some distance away from the rearrangement and is subject to a position effect. However, most known examples of position effects involve translocations or deletions rather than insertions (12).
Here, we report the complete genomic sequencing of the 85-kb triplication repeat unit and the identification of three genes wholly or partially encoded within it. The ubiquitination factor Ufd2 becomes linked to D4Cole1e and encodes a fusion protein, whereas a novel member of the cellular retinoid-binding protein family, Rbp7, is overexpressed in the WldS mouse. Of the two transcripts, only the chimeric mRNA is abundantly expressed in nervous tissue, suggesting it is a good candidate for slowing the rate of Wallerian degeneration. The identification of these gene alterations reveals that the WldS mutant could be useful for studies of ubiquitin and retinoid metabolism as well as axon degeneration.
Materials and Methods
Subcloning and Sequencing.
The identification of bacterial artificial chromosome (BAC) and P1 artificial chromosome (P1) clones has been reported previously (9). The BAC 269F8 (Research Genetics, Huntsville, AL) was partially digested with MboI and subcloned into LambdaGEM-12 XhoI half-site arms (Promega). Phage, P1 (320-I15, RZPD, Berlin), and BAC 269-F8 (directly) were then subcloned into pGEM3, pGEM7 (Promega), or pcDNAII (Invitrogen) according to the availability of suitable cloning sites. PCR products were generated using the high-fidelity polymerase Pfu (Stratagene) and cloned into pCR-Script (Stratagene). Those larger than 2.0 kb were first fragmented using blunt-end cutting restriction enzymes.
Automated DNA sequencing reactions were done using Amersham Thermosequenase Dye Primer and Dye Terminator kits. Sequencing reactions were analyzed at Alta Bioscience (University of Birmingham) using an ABI373a or 377 prism sequencer and the corresponding ABI software. Raw sequence was assembled into contigs using the GCG program GELASSEMBLE.
Exon Trapping.
Internal exon trapping was performed as previously described (13) using DNA from BAC 269F8 and BAC 271D15 (Research Genetics) and P1 320-I15,11L19 and P1 301N12 (RZPD, Berlin) subcloned using EcoRI plus BamHI or EcoRI plus PstI.
Sequence-Based Exon Prediction.
Genomic DNA sequence was used to search GenBank nonredundant and dbEST databases using both BLASTN and BLASTX (2.0.11) algorithms.
GRAIL 2 potential exons were identified using the ORNL GRAIL server (http://compbio.ornl.gov/Grail-1.3/), subjected to extensive analysis by BLASTN and BLASTX database searching and finally by RT-PCR (below).
Generation of Full-Length Transcripts.
RACE (3′-rapid amplification of cDNA ends) was performed to extend the identified exons in the 3′ direction, using the kit from GIBCO/BRL with modification previously described (13). Gene-specific custom primers were obtained from GIBCO/BRL.
The identified exons were also used as probes to screen a Lambda Zap II mouse brain cDNA library (ICR outbred strain, newborn, from Stratagene) by hybridization (14). The cDNA contig was extended by successive rescreenings of the cDNA library using the identified cDNA clones as probes.
RT-PCR was performed as described in ref. 13; total RNA was reverse transcribed using random primers and Superscript II reverse transcriptase (GIBCO/BRL). The first strand cDNA was then used for PCR using Pfu polymerase (Stratagene). RT-PCR products were cloned into pCR-Script Amp SK(+) (Stratagene) for sequence analysis.
The sequence of cDNA from WldS mice was compared with that from the closely related strain C57BL/6J. It is not known for certain that the WldS mutation arose on a C57BL/6J background, but it has been demonstrated previously that other C57 strains do not carry the triplication (9), so other strains are not expected to show the changes we describe below.
Northern Blotting Analysis.
Total RNA was isolated from 6–8-week-old C57BL/WldS and C57BL/6J mouse tissues using the single-step guanidinium thiocyanate method (15). Northern blotting was performed using 1.0% agarose/formaldehyde gels and blotting overnight onto Trans-blot nitrocellulose membrane (Bio-Rad) in DEPC-treated 10× SSC. Probes were labeled using the Megaprime kit (Amersham Pharmacia) and [α-32P]dCTP (Amersham Pharmacia) and hybridized for 1 h at 68°C in Expresshyb (CLONTECH). Posthybridization washes were done in 1× SSC, 0.1% SDS at 50°C, and the filters were exposed to x-ray film (Xograph) for 1–5 days at −80°C.
Generation of Polyclonal Antibodies to Synthetic Peptides.
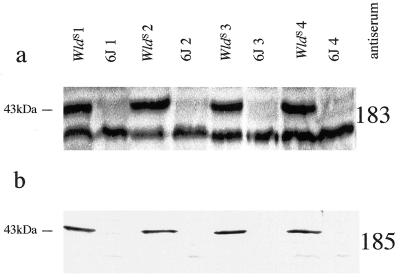
The predicted protein sequence of the Ufd2/D4Cole1e fusion protein was analyzed using the GCG program ANTIGENIC to predict the peptides most likely to provoke a strong antibody response upon immunization. Peptides were then chosen for custom synthesis (Research Genetics) based on their predicted antigenicity and their location in different parts of the primary structure. Peptide CIRYLVPDLVQEYIEK is located close to the C terminus and was used to generate antiserum 183. Peptide CTSPIGAADNIAVRGLH crosses the fusion boundary and was used to generate antiserum 185 (C indicates an additional cysteine added for conjugation purposes). Each peptide was coupled through its N-terminal cysteine to keyhole limpet hemocyanin (Sigma) using the heterobifunctional agent MBS (Sigma) as described (16) and immunized into two New Zealand White rabbits. After four immunizations at intervals of 2–4 weeks, the antisera reacted against both the corresponding peptide (data not shown) and a WldS-specific protein of the expected molecular weight (see Fig. 4). Some nonspecific bands were also observed with antiserum 183.
Figure 4.
Western blots of mouse brain homogenate with antisera 183 (a) against a peptide derived from C-terminal region sequence of D4Cole1e and 185 (b) against a peptide whose sequence spans the junction between Ufd2 and D4Cole1e. Each antiserum was used at 1:500 dilution and detects a consistently WldS-specific 43-kDa protein. Antiserum 183 also detects a 40-kDa protein, which may be either nonspecific or the endogenous D4Cole1e product. This additional band indicates similar loading and transfer of in the WldS and C57BL/6J lanes. Samples derived from different animals are distinguished by the labels WldS 1, WldS 2, etc.
Western Blotting Analysis.
Mouse brains were homogenized in twice the wet volume of 20 mM Hepes (pH 7.5), 0.2 M CaCl2, 0.2 M MgSO4, protease inhibitor mixture (1 ml per 20 g of tissue; Sigma), 1 mg/ml DNase (Sigma). Proteins were solubilized in a standard Laemmli sample buffer and separated by SDS/PAGE [12% (wt/vol) acrylamide] before transferring onto nitrocellulose using a semidry blotter (Bio-Rad). Ponceau S staining (Sigma) of blots was used to confirm approximately equal loading of lanes and successful transfer of protein. Blots were incubated with primary antibodies overnight at 4°C and with secondary antibody (goat anti-rabbit-horseradish peroxidase; Dianova, Hamburg, Germany) for 1 h at room temperature. Signals were detected by enhanced chemiluminescence (Amersham Pharmacia) according to the manufacturer's instructions.
Results
Sequence Analysis and Identification of Exons.
The sequence between the previously reported boundaries of the triplication repeat unit (9) was determined together with a few kilobases either side. Putative exons were identified from the completed sequence using BLASTX and BLASTN (2.0.11) and GRAIL (Fig. 1). Further putative exons from the repeat unit and the immediately surrounding region were identified by exon trapping of P1 301N12, P1 11L19, and BAC 269F8.
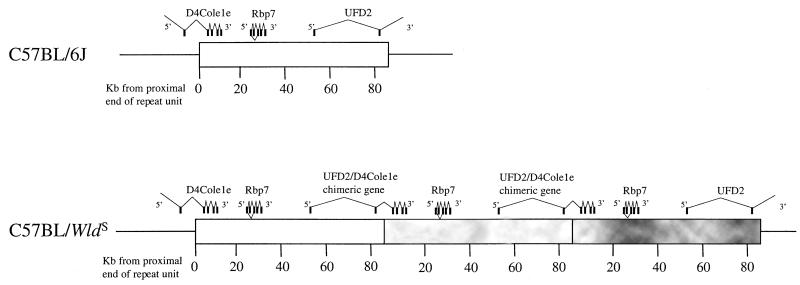
Figure 1.
Location of exons within the 85-kb WldS triplication repeat unit. In WldS, three adjacent repeat units are shown to indicate (a) how Ufd2 and D4Cole1e are brought together to form a chimeric gene, (b) that normal copies of Ufd2 and D4Cole1e exist still at either end of the repeat array (although the complete genes are not shown), and (c) how Rbp7 is present at three times the normal copy number.
Three clusters of exons were identified within the repeat unit. First, two exons located approximately 30 kb and 2.5 kb proximal to the distal end of the repeat unit were found to encode the 5′-UTR and N-terminal coding region of ubiquitin fusion degradation protein 2 (Ufd2), a protein required for the addition of multiubiquitin chains to a ubiquitinated substrate (17). The predicted amino acid sequence in this region is identical to the human Ufd2 sequence (GenBank accession no. AF043117) but represents an N-terminal extension of the yeast Ufd2 protein. Further exons of Ufd2 were identified by exon trapping of P1 clones distal to the repeat unit. Second, four exons 5–11 kb from the proximal end of the repeat unit were found to be homologous to three yeast genes (SwissProt accession nos. P53204 and S53405 and GenBank accession no. AL117212) and two Caenorhabditis elegans genes (F26H9.4 and AL02275) (all at 33–38% identity at the amino acid level). The yeast genes P53204 and S53405 are predicted to encode single-pass membrane-spanning proteins, but the mouse protein is not predicted to be membrane spanning by the TMPRED and TMAP algorithms. The sequence of the mouse gene, D4Cole1e, provides no further clues as to the function of the protein. Third, four exons and a fifth alternatively spliced exon located 24–30 kb from the proximal end of the repeat unit are homologous to the family of cellular retinol-binding proteins. The homology with murine CRBPI and CRBPII (18, 19) is particularly strong (57% and 56% amino acid identity, respectively). In view of this homology, the gene was named Rbp7 for retinoid-binding protein. Two additional potential exons identified by exon trapping (269ET37 and 11ET38) are spliced to the 5′ end of the second exon in 3′-RACE and RT-PCR products (data not shown). They are, however, completely absent from the EST database so it is not clear whether they make a significant contribution to Rbp7 transcripts.
All of these exons are located in the same chromosomal orientation (5′ proximal, 3′ distal). Therefore, RT-PCR and 3′-RACE using C57BL/WldS and C57BL/6J mouse brain first-strand cDNA was used to confirm that they are expressed and to determine which exons belong to which gene (data not shown). In normal mouse brain, the three clusters of exons were shown to derive from three independent genes as shown in Fig. 1.
Further cDNA library screening, 3′-RACE and RT-PCR, was used to determine the full coding sequence of murine Ufd2 and to extend D4Cole1e further in the 5′ direction. In both cases, the additional sequences lie outside the triplication unit.
Finally, an endogenous retroviral sequence was found to be located 40–46 kb from the proximal end of the repeat unit. There is no protein coding sequence in or in close proximity to it, so it is not clear whether it plays any functional role.
Identification of a Chimeric mRNA and Protein in WldS Central and Peripheral Nervous System.
In the WldS genome, Ufd2 and D4Cole1e are located on either side of the boundary between adjacent repeat units and lie in the same chromosomal orientation, so it was reasoned that they might splice together to form a chimeric transcript. RT-PCR using first-strand brain cDNA and one primer from each gene revealed the existence of such a chimeric transcript in WldS brain that is not present in control brain from C57BL/6J (Fig. 2a). This result was confirmed using a new primer, and cloning and sequencing of the Pfu PCR products confirmed that the cDNA is derived from both genes (Fig. 2d). The predicted protein is an in-frame fusion between the N-terminal 70 amino acids of Ufd2 and the C-terminal 302 amino acids of D4Cole1e along with an aspartic acid formed by the junction.
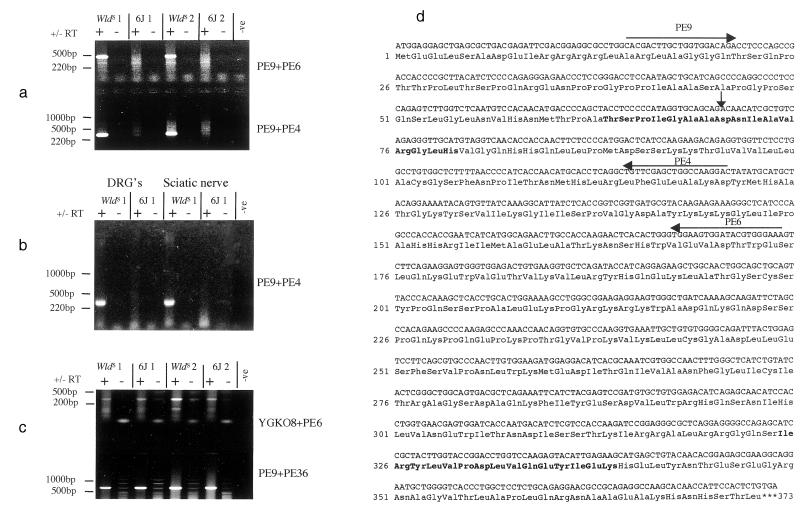
Figure 2.
(a) RT-PCR showing the detection in brain of a WldS-specific chimeric transcript between Ufd2 and D4Cole1e. Primers indicated in (d) were used to amplify fragments of 478 bp and 318 bp as shown. Samples derived from different animals are distinguished by the labels WldS 1, WldS 2, etc. (b) RT-PCR showing WldS-specific expression of the chimeric transcript in dorsal root ganglia (DRG) and sciatic nerve. (c) RT-PCR showing the expression of normal transcripts in both 6J and WldS brain for D4Cole1e (Upper) and Ufd2 (Lower) (expected sizes 334 and 710 bp, respectively). Primer sequences: YGK08 = 5′-ACTAGGGCCGTTTGGCTTC-3′; PE36 = 5′-CCTGAACTGGAGCCAGTGTT-3′; PE6 and PE9 as shown in (d). (d) Sequence of the chimeric cDNA and the predicted coding sequence. The vertical arrow indicates the junction between Ufd2-derived sequence and D4Cole1e-derived sequence (Asp-71 is derived from neither protein but formed by the junction). Horizontal arrows indicate the location, and 3′ direction of primers used in (a) and amino acids shown in bold are those used to make peptides for generation of polyclonal antisera 183 (Ile-325 to Lys-339) and 185 (Thr-64 to His-79).
Northern blotting using a 3′-D4Cole1e probe shows a WldS-specific band of approximately 3.2 kb, in good agreement with the size of the chimeric transcript predicted from the cDNA and genomic sequence (3.1 kb plus polyA tail) (Fig. 3a). The transcript is widely expressed, consistent with the observation that Ufd2 (whose promoter drives the expression) has EST in a wide variety of tissues. However, there is a particularly high level of expression in brain. The chimeric transcript was also detected in dorsal root ganglia and sciatic nerve by RT-PCR (Fig. 2b), indicating that it is expressed in the peripheral nervous system too.
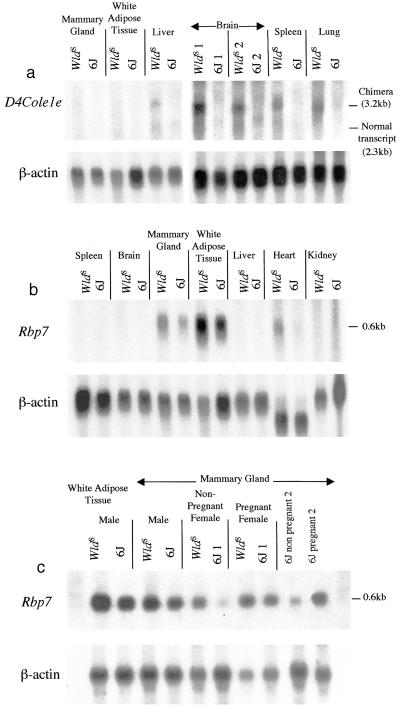
Figure 3.
(a) Northern blot showing WldS-specific expression of the chimeric transcript (as detected by a D4Cole1e-specific probe) and a high level of expression in brain. A faint band of 2.3 kb seen in liver in both strains probably corresponds to the normal D4Cole1e transcript. Samples derived from different animals are distinguished by the labels WldS 1, WldS 2, etc. A β-actin control indicates the quantity and integrity of RNA in each lane. (b) Northern blot showing expression of Rbp7 in tissues from C57BL/WldS and C57BL/6J mice. Expression is increased in WldS. (c) Northern blot showing higher expression of Rbp7 in male than in female (nonpregnant) mammary gland and up-regulation during pregnancy. The two right lanes show again the increase during pregnancy.
Due to the nature of the mutation, a single copy of each normal gene was expected to be present in WldS at the proximal or distal end, respectively, of the repeat array (Fig. 1). Expression of each normal gene was confirmed by RT-PCR (Fig. 2c), and a transcript of the expected size for the normal D4Cole1e is seen in Fig. 3a (liver). As expected, the expression level of these normal transcripts was unaltered between C57BL/6J and WldS brain.
Western blotting was used to confirm the existence and reading frame of the chimeric protein. Polyclonal antiserum 183 raised using a peptide whose sequence is located in the D4Cole1e part of the protein (Fig. 2d) bound to a protein of Mrapp 43 kDa, close to the predicted size of the chimeric protein (41 kDa), which was consistently present in WldS brain and absent from C57BL/6J brain (Fig. 4a). Similar data were obtained using a second polyclonal antiserum, 185, raised to a peptide whose sequence spans the junction between the two proteins (Fig. 4b). The fact that this protein is WldS-specific, has the expected Mr, and is recognized by antisera against two different parts of the predicted sequence indicates that it is the chimeric protein.
Overexpression of Rbp7 in WldS.
Northern blotting indicated that, of the tissues tested, Rbp7 is expressed predominantly in the mammary gland and white adipose tissue, with a lower level of expression in heart (Fig. 3b). The level of expression in mammary gland was lower in females than in males but consistently increased during pregnancy (Fig. 3c). No mRNA was detected in brain on Northern blots, but using RT-PCR expression of Rbp7 was detected in brain, dorsal root ganglia, sciatic nerve, and kidney.
Cloning and sequencing of the brain RT-PCR product confirmed that the sequence of the transcript is unaltered in WldS mice (data not shown). However, in WldS the level of expression of the normal-sized transcript is increased both in the tissues seen on the Northern blot and in brain, as shown by semiquantitative RT-PCR (data not shown).
Discussion
We have identified two transcripts that are directly altered by the WldS triplication. One is formed by splicing together across the triplication domain boundary of exons from the genes Ufd2 and D4Cole1e, and it encodes an in-frame fusion protein. The second transcript, Rbp7, is overexpressed with no alteration to its sequence. The expression patterns suggest that the Ufd2 fusion protein is the better candidate for Wld.
The presence of the N-terminal 70 amino acids of Ufd2 in the fusion protein is intriguing in view of the many reports of altered ubiquitin metabolism in neurological disease. Ubiquitinated deposits accumulate in Alzheimer's disease neuritic plaques (20), Parkinson's disease Lewy bodies (21), and in the intranuclear inclusion bodies of polyglutamine diseases Huntington's chorea (22) and SCA1 (23). In the axon, ubiquitin accumulates in four-repeat tau transgenic mice (24) and following spinal cord injury (25). These reports suggest that the ubiquitin proteolytic pathway plays an important role in axonal degeneration as well as in neurodegeneration in general. The recent discovery in the gracile axonal dystrophy (gad) mouse of an intragenic deletion in the ubiquitin carboxyl-terminal hydrolase gene UCH-L1 indicates that alterations to ubiquitin metabolism can play a causative role in axon degeneration (26).
The WldS mouse may be a useful system in which to investigate the role of Ufd2 regardless of whether the chimeric transcript causes slow Wallerian degeneration. For example, the fusion protein could sequestrate any protein that binds to the N-terminal 70 amino acids of Ufd2, and it would be interesting to investigate the consequences for the ubiquitin proteolytic pathway. Ufd2 is known to be required for the multiubiquitination of ubiquitinated substrates in yeast and requires also E1, E2, and E3 enzymes to function (17). Its disruption impairs cell survival in stress situations, which would be consistent with a role in prolonging the survival of an injured axon. However, apart from this, little is known about the function and mechanism of action of Ufd2. Its substrates are not known, and the only known binding protein in yeast is CDC48. The presence in mammals of a long N-terminal extension (including the 70 amino acids of the WldS-specific fusion protein) suggests that mammalian Ufd2 may have additional properties.
Similarly, studies in WldS could reveal something of the function of D4Cole1e. The C-terminal region of this protein is present in the WldS-specific fusion protein, and, compared with wild-type D4Cole1e, the chimeric transcript is heavily overexpressed in WldS brain. This overexpression may result both from the strength of the Ufd2 promoter in brain and the presence of two copies of the chimeric gene per haploid genome. Almost nothing is known of the normal function of D4Cole1e, although its higher level of expression in liver than in other tissues might indicate a role in this organ. The D4Cole1e part of the WldS-specific fusion protein could also play an important role in the slow Wallerian degeneration phenotype, for example through a gain-of-function due to loss of a regulatory domain or fusion with the new sequences.
We also report the identification of a new member of the retinoid-binding protein family, Rbp7, expressed predominantly in white adipose tissue and mammary gland. Its homology to other members of the cellular retinoid-binding protein family, and to retinol-binding proteins in particular (Fig. 5), suggests that Rbp7 is likely to function in retinol sequestration and/or metabolism in these tissues. The increase during pregnancy suggests that one function of Rbp7 could be the storage of retinol in mammary gland and/or secretion of retinol in milk. It is not clear why expression in nonpregnant females should be lower than in the male mammary gland. Although it is conceivable that an alteration in retinoid metabolism could influence axonal survival, given that retinoic acid can induce neuronal differentiation (27), the very low level of expression of Rbp7 in brain would seem to argue against such a role. However, the demonstration that the transcript is overexpressed in WldS indicates that it is important to look for alterations to retinoid metabolism in this mutant. Like the CRBPI-overexpressing transgenic (28), the WldS mouse shows no overt signs of vitamin A toxicity or deficiency, but it has not been studied in the context of vitamin A-enriched or -deficient diets. Such diets were necessary to reveal an abnormal phenotype in the CRBP1 null mutant (29). A further possibility is that up-regulation of Rbp7 could play a compensatory role in the CRBPI null mutant.
Figure 5.
Alignment of amino acid sequences of retinoid-binding protein family members and peripheral myelin protein P2, showing the close relationship of Rbp7 with CRBPI and CRBPII. CRABPI, CRABPII, and P2 diverge from the CRBP family particularly at their C-terminal ends. Amino acids that are totally conserved in these proteins are shown in bold, and the percentage identity with Rbp7 is shown at the end. A dash indicates an identical amino acid to Rbp7; periods indicate a gap introduced to optimize the fit.
In summary, we report the identification of two transcripts that are directly altered by the tandem triplication in the slow Wallerian degeneration mouse. The fusion protein formed from Ufd2 and D4Cole1e is a good candidate because of its strong and widespread expression in the nervous system. Rbp7, whose normal transcript is overexpressed in WldS mice, seems to function in mammary gland and white adipose tissue. These data indicate both a candidate gene for Wld and the importance of studying ubiquitin and retinoid metabolism in the WldS mouse.
Acknowledgments
We are grateful to Dr. Paul Edwards (University of Cambridge) for technical advice and helpful discussion and to Dr. Trevor Dale (Imperial Cancer Research Fund, London) for technical advice. These studies were funded by Bristol-Myers Squibb, Inc. and by the Center for Molecular Medicine of the University of Cologne (ZMMK). L.C. is a fellow of the Mario Negri Pharmacological Research Institute (Milan). The P1 clone used for some of the sequencing was provided by the Resource Center Primary Database (RZPD), Berlin.
Abbreviations
- WldS
slow Wallerian degeneration mutant
- P1
P1 artificial chromosome
- BAC
bacterial artificial chromosome
- RACE
rapid amplification of cDNA ends
- kb
kilobase(s)
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF260923 (Mouse Rbp7 Complete cds), AF260924 (WldS-specific chimeric cDNA complete cds), AF260925 (D4Cole1e Partial cds), AF260926 (Mouse Ufd2 Complete cds), and AF260927 (WldS triplication genomic sequence)].
References
- 1.Lunn E R, Perry V H, Brown M C, Rosen H, Gordon S. Eur J Neurosci. 1989;1:27–33. doi: 10.1111/j.1460-9568.1989.tb00771.x. [DOI] [PubMed] [Google Scholar]
- 2.Waller A. Philos Trans R Soc London. 1850;140:423–429. [Google Scholar]
- 3.Dal Canto M C, Gurney M E. Brain Res. 1995;676:25–40. doi: 10.1016/0006-8993(95)00063-v. [DOI] [PubMed] [Google Scholar]
- 4.Fujimura H, Lacroix C, Said G. Brain. 1991;114:1929–1942. doi: 10.1093/brain/114.4.1929. [DOI] [PubMed] [Google Scholar]
- 5.Buckmaster E A, Perry V H, Brown M C. Eur J Neurosci. 1995;7:1596–1602. doi: 10.1111/j.1460-9568.1995.tb01155.x. [DOI] [PubMed] [Google Scholar]
- 6.Finn J T, Weil M, Archer F, Siman R, Srinivasan A, Raff M C. J Neurosci. 2000;20:1333–1341. doi: 10.1523/JNEUROSCI.20-04-01333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernier B, Castejon S, Culver D G, Glass J D. NeuroReport. 1999;10:1423–1426. doi: 10.1097/00001756-199905140-00007. [DOI] [PubMed] [Google Scholar]
- 8.Lyon M F, Ogunkolade B W, Brown M C, Atherton D J, Perry V H. Proc Natl Acad Sci USA. 1993;90:9717–9720. doi: 10.1073/pnas.90.20.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman M P, Conforti L, Buckmaster E A, Tarlton A, Ewing R M, Brown M C, Lyon M F, Perry V H. Proc Natl Acad Sci USA. 1998;95:9985–9990. doi: 10.1073/pnas.95.17.9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmerman V, Nelis E, Van Hul W, Nieuwenhuijsen B W, Chen K L, Wang S, Ben Othmane K, Cullen B, Leach R J, Hanemann C O, et al. Nat Genet. 1992;1:171–175. doi: 10.1038/ng0692-171. [DOI] [PubMed] [Google Scholar]
- 11.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. Nature (London) 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 12.Kleinjan D J, van Heyningen V. Hum Mol Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 13.Conforti L, Buckmaster E A, Tarlton A, Brown M C, Lyon M F, Perry V H, Coleman M P. Mamm Genome. 1999;10:617–622. doi: 10.1007/s003359901056. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Chomczynski P, Sacchi N. Anal Biochem. 1978;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Liu F-T, Zinnecker M, Hamaoka T, Katz D H. Biochemistry. 1979;18:690–697. doi: 10.1021/bi00571a022. [DOI] [PubMed] [Google Scholar]
- 17.Koegl M, Hoppe T, Schlenker S, Ulrich H D, Mayer T U, Jentsch S. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 18.Smith W C, Nakshatri H, Leroy P, Rees J, Chambon P. EMBO J. 1991;10:2223–2230. doi: 10.1002/j.1460-2075.1991.tb07758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakshatri H, Chambon P. J Biol Chem. 1994;269:890–902. [PubMed] [Google Scholar]
- 20.Mori H, Kondo J, Ihara Y. Science. 1987;235:1641–1644. doi: 10.1126/science.3029875. [DOI] [PubMed] [Google Scholar]
- 21.Li K, Ito H, Tanaka K, Hirano A. J Neuropathol Exp Neurol. 1997;56:125–131. doi: 10.1097/00005072-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 22.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 23.Cummings C J, Mancini M A, Antalffy B, DeFranco D B, Orr H T, Zoghbi H Y. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 24.Spittaels K, Van den Haute C, Van Dorpe J, Bruynseels K, Vandezande K, Laenen I, Geerts H, Mercken M, Sciot R, Van Lommel A, et al. Am J Pathol. 1999;155:2153–2165. doi: 10.1016/S0002-9440(10)65533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G L, Farooque M. Acta Neuropathol. 1996;91:155–160. doi: 10.1007/s004010050407. [DOI] [PubMed] [Google Scholar]
- 26.Saigoh K, Wang Y L, Suh J G, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, et al. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 27.Sidell N, Altman A, Haussler M R, Seeger R C. Exp Cell Res. 1983;148:21–30. doi: 10.1016/0014-4827(83)90184-2. [DOI] [PubMed] [Google Scholar]
- 28.Troen G, Eskild W, Fromm S H, De Luca L M, Ong D E, Wardlaw S A, Reppe S, Blomhoff R. J Nutr. 1999;129:1621–1627. doi: 10.1093/jn/129.9.1621. [DOI] [PubMed] [Google Scholar]
- 29.Ghyselinck N B, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson C B, Hakansson H, Sauvant P, et al. EMBO J. 1999;18:4903–4914. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]