Abstract
We performed a gene expression screen of the entire transcriptome of the major African malaria vector Anopheles gambiae for immune response genes in adult female mosquitoes, which is the developmental stage infected by malaria parasites. Mosquitoes were immune-stimulated for subtractive cloning by treatment with bacterial lipopolysaccharide, a potent and general elicitor of the innate immune response, and by injury. The screen yielded a highly enriched cDNA library in which more than half of the clones were immune responsive. In this paper, we describe 23 immune-regulated genes, including putative protease inhibitors, serine proteases, regulatory molecules, and a number of genes without known relatives. A molecule related to the protease inhibitor α-2-macroglobulin responded strongly to malaria parasite infection, but displayed little or no response to bacteria, whereas other genes exhibited the inverse pattern. These results indicate that the insect immune system discriminates between molecular signals specific to infection with bacteria and malaria parasites.
Malaria remains one of the major infectious diseases of the world, affecting the health of hundreds of millions of people. A series of molecular and cellular interactions occur during passage of the parasite through the mosquito vector, which may offer opportunities for development of mosquito-based molecular strategies to interrupt malaria transmission (1, 2). Parasite development within the vector begins when the mosquito takes an infective bloodmeal from a vertebrate host. Sexual fertilization occurs within the blood in the mosquito midgut, and the resulting zygotes transform to motile ookinetes. About 24 h after the bloodmeal, the resulting ookinetes invade the epithelial cell layer of the midgut, where they transform to oocysts. Each oocyst undergoes numerous internal mitotic divisions to yield thousands of sporozoites that are released into the mosquito body cavity. When sporozoites invade the salivary glands, about 10 days after the initial infective bloodmeal, the mosquito becomes competent to transmit malaria.
There are measurable costs to malaria-infected mosquitoes in components of reproductive fitness such as longevity, fecundity, and flight distance (3–7), which could drive the natural selection of antiparasite immune surveillance and effector functions. In fact, malaria parasites suffer large numerical losses during development within the mosquito, and ultimately only a small minority of the parasites that enter the vector develop completely (8, 9). At least some of these losses probably result from vector immune mechanisms that attenuate the efficiency of parasite development. Factors induced during infection, including nitric oxide, could play such a role (10, 11). In addition, two described genetic mechanisms can render anophelines nonpermissive for Plasmodium (12, 13). Nevertheless, the numbers of surviving, successfully developing parasites in the vector are at least adequate to maintain disease transmission in nature.
Molecular features of the insect immune response to bacteria have shown that the insect immune system is clearly the phylogenetic precursor of the innate immune system of vertebrates (14, 15). The mechanisms involved in the insect immune response to eukaryotic microbial pathogens have been examined only recently, using mosquitoes infected with malaria parasites as a model. To date, the following genes have been found to respond transcriptionally in malaria-infected mosquitoes: the antibacterial peptide defensin, a Gram-negative bacteria-binding-related protein (GNBP), a chitin-binding-related protein (IGALE20) (16, 17), nitric oxide synthase (11, 18), and three serine protease relatives (19).
These parasite-responsive genes also responded to bacterial infection, and thus did not suggest pathogen-specific immune signaling. Fungi and bacteria induce distinct immune signaling pathways in Drosophila and cause differential transcriptional responses of antimicrobial peptides (20, 21). Drosophila immune gene expression in response to protozoan or other eukaryotic pathogens has not been examined in detail, although genetic studies identified a locus that seems to control response to parasitoid wasp eggs independently of the antibacterial response (22).
The previously described mosquito immune genes were identified by degenerate PCR (11, 19) or differential display (23). These approaches both require identification and cloning of individual gene fragments. Here, we present a global gene expression screen for immune response genes in adult female Anopheles gambiae mosquitoes. The screen used subtractive enrichment by solution hybridization and yielded a highly enriched immune-response library. Expression analysis revealed both overlapping and distinct genetic programs of immune gene regulation in response to infection with bacteria and malaria parasites, and at least one gene responded more to malaria parasites than to bacteria. The resulting enriched library is suitable in quality and complexity for cDNA microarray production and functional genomic analysis of mosquito immune response.
Materials and Methods
Mosquitoes and Parasites.
A. gambiae (G3 strain) mosquitoes were reared at 27 ± 1°C and 85 ± 5% relative humidity with 12-h cycles of alternating light and darkness. Mosquitoes were offered 20% Karo sugar syrup on cotton pads for routine maintenance. Malaria parasites were Plasmodium berghei NK-65 strain. The gametocyte nonproducing 2.33 strain, which infects erythrocytes but is noninfective to mosquitoes (24), was used as a control.
Immune Stimulation, mRNA, and cDNA.
For immune stimulation, adult female mosquitoes 2–4 days old were inoculated with bacterial lipopolysaccharide (LPS) from Escherichia coli 055:B5 (Sigma) in PBS at 1 mg/ml. Mosquitoes were anesthetized with CO2, and approximately 300 nl of LPS per mosquito was inoculated intrathoracically through a glass needle. Mosquitoes were returned to the insectary for 6 h. Control mosquitoes were anesthetized but not inoculated.
mRNA was purified from 20 each LPS-induced and control mosquitoes. Mosquitoes were decapitated to eliminate brain mRNAs, which are unlikely to be important in immune response. One microgram of LPS-induced and control RNA was used as substrate for oligo(dT)-primed synthesis of double-stranded cDNA. An aliquot of each cDNA was used for cDNA subtraction, as described below.
cDNA Subtraction.
Subtraction was done by iterative solution hybridization with PCR-amplified cDNA restriction fragments (25–27). Modifications from previous protocols included the development of a simple spin-cartridge method for tester enrichment after solution hybridization and the use of 40% mass amount of the original, unenriched cDNA as driver in each hybridization to suppress the reemergence of residual shared sequences, which eliminated the need for “short hybridization” steps used previously (26). A detailed protocol is available by request to K.D.V.
Briefly, double-stranded cDNAs were restricted with AluI and RsaI to maximize the probability of including cDNAs in the amplifiable pool, were ligated to a blunt-ended adapter made of annealed 5′-CTCTTGCTTGAATTCGGACTA-3′ and 5′-pTAGTCCGAATTCAAGCAAGAGCACA-3′, and were amplified by PCR using the nonphosphorylated oligo as primer. All steps were identically applied to LPS and control samples. LPS-treated driver cDNA was restricted with EcoRI to remove the ligated adapter and was labeled with photobiotin (28). Control tester was not cleaved or labeled.
Two parallel series of subtractions yielded libraries of LPS up-regulated and down-regulated genes, respectively. The initial solution hybridization reaction contained 25 μg of driver cDNA and 1.3 μg of tester. In each subsequent cycle, driver was 15 μg of the opposite enriched cDNA generated in the previous cycle plus 10 μg of the opposite unenriched cDNA from the beginning of the procedure. Solution hybridization reactions were denatured and then reassociated at 68°C overnight in buffer (27).
Unhybridized tester molecules were recovered by first using a negative selection to remove driver, including driver-tester heteroduplexes. Streptavidin was added to bind to biotin on the driver molecules, and conjugates were removed by Advamax protein-binding beads (Edge Biosystems, Gaithersburg, MD). Under these conditions, approximately 90% of biotinylated DNA molecules were removed whereas the same proportion of nonbiotinylated single-stranded cDNA remained in the supernatant. Next, the remaining single-stranded cDNA molecules were recovered in a positive selection by binding to a microfuge spin filter (AGTC Protein Cartridge; Edge Biosystems). After washing and elution, we recovered approximately 95% of single-stranded nonbiotinylated DNA molecules from the post-Advamax supernatant. Finally, the mass amount of enriched tester cDNA was amplified by PCR. After five cycles of subtraction, the LPS up-regulated enriched cDNA was cloned into plasmid vector PCR-Script (Stratagene).
Enriched Library Screen.
Randomly picked colonies from the LPS up-regulated enriched library were used as probes for blots of (i) 2 μg of EcoRI-digested A. gambiae genomic DNA, to verify that the cloned sequence came from the mosquito genome, and (ii) 1 μg each of control and LPS unenriched cDNA from the beginning of the procedure, to verify that the sequence was immune-regulated.
Virtual Northern Blots.
Virtual Northern blots were used to assess mRNA expression levels for individual genes, circumventing the problem of measuring mRNAs in limiting amounts of biological material. Virtual Northern blots were blots of full-length cDNA, made by PCR amplification of the primary cDNA (29–32). We found consistent reproduction of total cDNA populations for molecules up to ≈4 kb.
Double-stranded cDNA made as described above was ligated to adapters 5′-AATTCGCGGCCGCGTCGAC-3′ and 5′-pGTCGACGCGGCCGCG-3′. Five nanograms of cDNA was used as PCR template with the nonphosphorylated adapter oligo as primer and Taq Advantage cDNA enzyme (CLONTECH) for 25 cycles with annealing temperature 60°C. Eight nanograms of reaction products then were used as template in a new reaction of seven cycles with the same parameters.
cDNA (150 ng), which was an image of the poly(A)+ mRNA population, was separated by electrophoresis and transferred to a nylon membrane. After hybridization to experimental probes, blots were stripped and reprobed with A. gambiae ribosomal protein rpS7 as an internal loading control (33, 34). Blots were imaged with a PhosphorImager SI (Molecular Dynamics) and signals were quantified by using imagequant software. A correction factor based on rpS7 signal in each lane controlled for loading variation between lanes. All graphed values or comparisons of expression levels are based on corrected signals.
Sequence Analysis.
Sequence homology searches of public databases were carried out with the blast programs with the default BLOSUM-62 substitution matrix (35). Homologies were considered statistically significant if a blastp search of the nonredundant peptide database with a plausible ORF from the query sequence generated an Expect (E) value <0.05 (35). Sequence alignments of protease relatives were done with clustalw Version 1.8. Additional sequence analysis was carried out by using programs in the dnastar (Lasergene, Madison WI) and gcg (GCG) program suites.
Immune Assays.
Adult mosquitoes 2–4 days old were subjected to the following treatments. To suppress native flora, a solution of 100 units/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml gentamycin, and 2,000 units/ml antifungal nystatin in 20% Karo syrup was exclusively offered on cotton pads for 3 days. For bacterial response, mosquitoes were inoculated intrathoracically with LPS as described above, or the lateral thorax was pierced with a stainless steel minuten pin (0.15-mm diameter; Bioquip Products, Gardena, CA) that was dipped into an equal-volume mixture of overnight LB cultures of Gram-negative E. coli 1106 (American Type Culture Collection no. 35581) and Gram-positive Micrococcus luteus (American Type Culture Collection no. 4698). For injury controls, mosquitoes were inoculated with sterile-filtered PBS, or the lateral thorax was wounded with a minuten pin. Wounds were not necessarily sterile, because microflora from the surface of the cuticle could have been introduced by the injury. For malaria infection, mosquitoes were fed on a hamster infected with P. berghei NK65 and were maintained at 21°C to allow parasite development. For normal bloodmeal, the hamster was not infected. For controls with infected but noninfective blood, mosquitoes were fed on P. berghei strain 2.33.
Mosquitoes were anesthetized with CO2, and RNA was prepared immediately. Northern blots were hybridized sequentially with experimental probes and with control rpS7. Blots were detected and quantified by PhosphorImager, and all experimental signals were corrected for loading variation by using rpS7 signal.
Results
Enrichment.
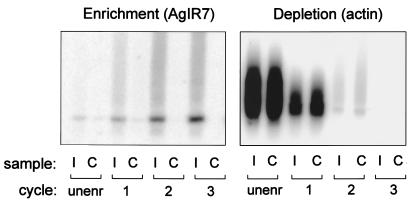
One microgram each of control and LPS-treated cDNA from the unenriched population and products of subtraction cycles, hybridized with clone AgIR7 from the LPS up-regulated library, showed progressive enrichment of this differentially expressed sequence (Fig. 1A). To detect depletion of shared sequences, the blot was hybridized to a probe for cytoplasmic actin (Fig. 1B and ref. 36), which was expressed at a high level in both unenriched samples. Actin signal decreased with each subtraction cycle, concomitantly with the enrichment of differential sequences such as AgIR7, and was undetectable by subtraction cycle 3.
Figure 1.
Enrichment of immune response genes by solution hybridization cycles. One microgram each of unenriched starting cDNA and products of sequential enrichment cycles were Southern blotted and probed with a differentially expressed sequence (AgIR7) (Left) or a sequence that was initially shared between the samples (cytoplasmic actin) (Right). Samples: cDNA from mosquitoes immune-stimulated by inoculation of LPS (I); control cDNA from untreated mosquitoes (C). Cycles: unsubtracted cDNA (unenr); cDNA after indicated number of subtraction cycles (cycles 1–3).
Screening.
To evaluate the efficiency of enrichment, 32 clones were randomly picked from the LPS up-regulated enriched library (the LPS down-regulated library was not examined). The clones were screened by hybridizing to genomic DNA and unenriched cDNA. We found that 59% (19/32) of randomly picked clones hybridized differentially to the unenriched cDNAs, indicating that they were regulated by immune stimulation. Of the 32 clones, 9 did not hybridize detectably to genomic DNA and/or to cDNA. They were either artifacts or rare mRNAs in the total population and were not characterized further. Considering only the clones that gave a signal in the primary screen, 83% (19/23) were immune-regulated.
Clones that survived the primary screen were subjected to a secondary screen by hybridizing to virtual Northern blots of full-length cDNA from control and LPS-treated mosquitoes. This step provided independent verification of gene regulation during immune response and indicated mRNA size. Representative virtual Northern results are shown in Fig. 2. The relevant bands are shown, and there were not significant secondary bands. As a control, mRNA from the rpS7 gene, which is not immune-regulated, displayed equivalent abundance between control and LPS-treated samples (Fig. 2), thus behaving the same as on standard Northern blots (Figs. 3–5), and in reverse transcription-PCR assays (23). We additionally verified for six genes that virtual and standard Northern blots gave the same size and expression profile (data not shown).
Figure 2.
Virtual Northern blots of immune-regulated genes. Probes for the genes indicated were hybridized to full-length cDNA from mosquitoes immune-stimulated by LPS (Imm) or untreated control mosquitoes (Ctrl). Blots showed the expression level of immune-stimulated mRNA as compared with controls, and the size of cognate mRNAs determined by comparison to size standards. Up-regulation levels and sizes are listed in Table 1. (Bottom) Sample was probed with the unregulated loading control ribosomal protein rpS7, mRNA size 0.7 kb.
Figure 3.
Immune regulation of serine protease relative AgISPR5. Mosquitoes were subjected to the indicated experimental treatment, and a Northern blot of poly(A)+ RNA from 20 mosquitoes per lane was probed with AgISPR5. Samples were untreated (Control), 3-day antibiotic treatment (Antibiotic), 6 h after inoculation with sterile saline (Injury), 6 h after inoculation with LPS (LPS), 24 h after taking a normal uninfected bloodmeal (Normal), and 24 h after a P. berghei-infective bloodmeal (Infective). (Inset) Northern blot lanes in the same order probed with AgISPR5 (Upper; 1.9 kb) and rpS7 loading control (Lower; 0.7 kb).
Figure 5.
Immune regulation of putative protease inhibitor AgIMcr14. Northern blot of total RNA isolated from 15 mosquitoes per lane after the indicated experimental treatment, probed with AgIMcr14. Mosquitoes were untreated (Control), 6 h and 24 h after thoracic wounding with a sterile needle (Injury), 6 h and 24 h after thoracic wounding with bacterial culture (Bacteria), 24 h after taking a normal uninfected bloodmeal (Normal), and 24 h after a P. berghei-infective bloodmeal (Infective). (Inset) Northern blot lanes in the same order probed with AgIMcr14 (Top; 4.4 kb), loading control rpS7 (Middle; 0.7 kb), and AgIR28 (Bottom; 1.4 kb).
Immune Response Gene Identities.
Twenty-three immune-regulated genes were identified in the enriched library by the screens described above and by other experiments. The genes and their putative functional identities are summarized in Table 1. New genes were designated AgI, for “A. gambiae immune-responsive,” followed by a name if the homology was significant (E < 0.05) or otherwise by a number. Two genes recovered from the enriched library were previously described as immune genes in A. gambiae and were not included in the table, a serine protease-related gene ISPL5 (GenBank accession no. AJ000675) and lysozyme (GenBank accession no. U28809). Only two of the genes described here were found more than once among the randomly picked clones, indicating that there is considerable complexity in the enriched library.
Table 1.
Immune-responsive A. gambiae genes, isolated from enriched cDNA library
| Clone | -Fold upregulated* | mRNA size, kb* | cDNA sequence | Sequence homologies† | GenBank accession no. |
|---|---|---|---|---|---|
| AgISerpF1 | 4 | 1.8 | Frag | Protease inhibitor, serpin family, E = e–21 | AF203339 |
| AgIMcr14 | 5‡ | 4.4‡ | Frag | Protease inhibitor, α2-macroglobulin and complement C3, E = e–9 | AF203333 |
| AgISPR1 | 30 | 1.4 | Frag | Serine protease, chymotrypsin-like, E = e–26 | AF203336 |
| AgISPR5 | 13 | 1.9 | Full | Clip-domain serine protease, blood coagulation factor X, E = e–14 | AF203334 |
| AgISPR9 | 3 | 2.2 | Frag | Serine protease, Limulus hemocyte factor D, E = e–21 | AF203337 |
| AgISPR10 | 100 | 2.5 | Frag | Serine protease, trypsin-like, E = e–2 | AF203338 |
| AgISPR20 | 10 | 1.5 | Frag | Serine protease, Limulus hemocyte factor D, E = e–5 | AF203335 |
| AgIR2 | 2 | 1.5 | Frag | None | AF283260 |
| AgIR4 | 5 | 1.4 | Full | None | AF283261 |
| AgIR6 | 8 | ND | Frag | (DNA damage-induced protein) | AF283262 |
| AgIR7 | 4 | 1.1 | Full | p10/OS, E = e–37 | AF283263 |
| AgIR15 | 19 | 1.9 | Frag | Nuclear scaffold attachment factor A, E = e–1 | AF283264 |
| AgIHEXB | 3 | 1.9 | Frag | β-hexosaminidase, β chain, E = e–11 | AF283265 |
| AgIR28 | 100 | 1.4 | Full | (DNA polymerase/homeodomain protein) | AF283266 |
| AgIR29 | 5 | 0.7 | Frag | None | AF283267 |
| AgIrpS18 | 1.5 | 1.0 | Frag | Ribosomal protein S18, E = e–6 | AF283268 |
| AgIrpS26 | 4 | 0.8 | Frag | Ribosomal protein S26, E = e–13 | AF283269 |
| AgIBB1 | 7 | 1.8 | Frag | None | AF283270 |
| AgICC4 | 6 | 2.2 | Frag | None | AF283271 |
| AgICE1 | 2 | 1.5 | Frag | None | AF283272 |
| AgIPAH | 9 | 1.8 | Frag | Phenylalanine hydroxylase, E = e–11 | AF283273 |
| AgIDB12 | 12 | 0.7 | Frag | A. gambiae ce5, E = e–20; (anti-thrombin) | AF283274 |
| AgIsHSP | 7 | 0.7 | Frag | Small heat shock protein, hsp20 family, E = e–31 | AF283275 |
Full, full length cDNA sequence; Frag, cDNA fragment, 300–600 bp; ND, no data.
Based on virtual Northern blots from LPS-inoculated A. gambiae, except where indicated.
Based on blast homology comparisons. E indicates the order of magnitude of the best “Expect” value from a blastp sequence comparison to the nonredundant peptide database, where scores <0.05 are considered significant. Entries in parentheses were weaker and speculative.
Based on Northern blots from mRNA of bacteria-inoculated A. gambiae.
There were three protease inhibitor relatives. First, AgISerpF1 is related to serpin-family protease inhibitors. Serpins are involved in the regulation of innate immunity in mammals and insects (37, 38). AgISerpF1 displays homology to Drosophila serpin Spn43Ac, which is required for controlled signaling via the Toll receptor pathway (39). In the absence of Spn43Ac, expression of Toll-regulated immune peptides was constitutive.
Second, AgIMcr14 is related to mammalian α2-macroglobulin and complement C3 and represents the first member of this class of protease inhibitors described in an insect (40). The only similar arthropod gene, α2-macroglobulin from horseshoe crab, produced mRNA of similar size to AgIMcr14 (41). The sequenced portion of AgIMcr14 included the bait region that binds to target proteases (amino acids 1–49 based on horseshoe crab domains; ref. 42), but ended above the residues involved in thiolester bond formation with the captured proteases.
Third, AgIDB12 displayed significant local homology to an antithrombin isolated from mosquito salivary glands (45). Gene AgIDB12 was the same as an anonymous A. gambiae expressed sequence tag fragment (46).
At least five AgI serine protease-related genes were identified: AgISPR1, AgISPR5, AgISPR9, AgISPR10, and AgISPR20. These genes represent members of different functional families based on sequence homology and were different from A. gambiae protease-related genes described recently (19). Protease signaling cascades are involved in control of multiple facets of innate immunity, including complement activation in mammals, Toll-like receptor activity and LPS response in mammals and insects, and activation of the important invertebrate immune effector, phenoloxidase (15, 47–49). Full-length cDNA sequence for the 1.9-kb AgISPR5 revealed an exceptionally long N-terminal domain of 294 aa with a clip-like domain (50) and a C-terminal region that was strongly related to the serine protease catalytic domain. The AgISPR5 C-terminal domain lacked the canonical active site catalytic triad (H-D-S), an activation cleavage site (IVGG), and the conserved amino acids that define the specificity pocket. However, AgISPR5 had a potential hydrophobic signal sequence, and the six conserved cysteines found in other serine proteases were all present in the C-terminal domain of AgISPR5. Thus, the molecule is structurally highly conserved but probably does not display protease activity. A serine protease homolog of AgISPR5 in the horseshoe crab, factor D, also lacked the protease catalytic triad, but displayed autonomous antimicrobial activity against Gram-negative bacteria (51). The previously described A. gambiae ISPL5 also had a long N-terminal clip-like domain and probably no active site (52).
For the other four protease-related genes, the sequenced portions included the region of the active sites H and D, but did not extend to the region of the active site S. Sequence alignments showed that at least AgISPR1 and AgISPR9 possess active site H and D residues and the conserved cysteines, and thus may be enzymatically active (data not shown). AgISPR10 has the active site D and conserved cysteines but not the active site H, although the protease family homology of this molecule is more distant. AgISPR20 lacks active site H and D and the conserved cysteine residues and also displays distant homology to proteases. The function of these immune-regulated protease relatives remains to be determined.
AgIR7 displayed strong homology to a protein that was associated with tissue regeneration in the cockroach (53, 54). The function of the molecule in regeneration is not known, and the p10 protein has not been previously described as an immune-related molecule.
Homologies for two molecules suggest possible regulatory functions. AgIR15 is homologous to vertebrate scaffold attachment factor A (SAF-A), which binds to chromatin during gene expression (55), and its up-regulation may be caused by new transcriptional demands during the immune response. Relatives of this molecule have not been previously described in invertebrates. For the other gene, the highly immune-responsive AgIR28 full-length peptide sequence had short homologies to several DNA polymerases (although AgIR28 is too small to be a polymerase) and to the Xenopus homeodomain transcription factor Pitx2.
AgIHEXB displays strong homology to the β-hexosaminidase β chain. This lysosomally restricted acid hydrolase has not been previously described as immune-responsive. AgIHEXB up-regulation could facilitate lysosomal degradation either of infection-related macromolecules internalized via the endosomal pathway or of pathogens internalized by phagosomes.
AgIPAH is highly homologous to phenylalanine hydroxylase (PAH), which among insects has to date only been identified in Drosophila. PAH has not been previously described as an immune-related molecule. The immune regulation of AgIPAH in mosquitoes is probably related to the function of the phenoloxidase cascade, which is an integral component of invertebrate immunity. Activated phenoloxidase produces melanin, a component of immune capsules, with biosynthetic intermediates that display direct antibiotic activities (12, 56, 57). The rate-limiting substrate for melanin synthesis in insects is tyrosine, and hydroxylation of phenylalanine by PAH is the sole pathway for tyrosine biosynthesis (58, 59).
AgIsHSP is highly homologous to members of the Hsp20 small heat shock protein (Hsp) family. In general, Hsp genes are up-regulated by a variety of pathophysiological states, and have been described as LPS-response genes in other organisms (60, 61). In particular, the small Hsp proteins can protect cells from an apoptotic outcome after oxidative stress by decreasing intracellular concentration of reactive oxygen species (62, 63), which are known to be released during the insect immune response (11, 57).
Two genes, AgIrpS18 and AgIrpS26, were highly homologous to ribosomal proteins S18 and S26, respectively. The immune regulation of these genes could result from translational demands during the immune response. However, recent work reported antibiotic activity of peptide fragments derived from ribosomal proteins and suggested the intriguing possibility that antibacterial peptides evolved from such ribosomal proteins (64, 65). Other ribosomal protein genes are not immune-regulated (33), making the regulation of these genes potentially interesting.
Finally, many of the cDNAs had no significant relatives in the public databases, including cDNAs with full-length sequence information, and their identity as well as function remains to be determined.
Gene Regulation.
Functional Northern blot analysis revealed both overlapping and distinct programs of gene expression in response to immune signals. The serine protease-related AgISPR5 displayed a background level of steady-state mRNA in untreated mosquitoes (Fig. 3). The repression of this background level by antibiotic treatment of mosquitoes indicates that it was caused by environmental exposure to bacteria, rather than by basal promoter activity. AgISPR5 mRNA was increased by LPS treatment more than 5-fold over saline wounding controls and more than 10-fold over untreated mosquitoes. AgISPR5 expression also was induced during the period of ookinete invasion of the midgut 24 h after an infective bloodmeal, more than 2-fold in comparison to mosquitoes that received a normal bloodmeal. The mRNA response seen 24 h after a control normal bloodmeal was probably triggered by bacterial growth, because native midgut bacteria proliferate after a bloodmeal (11, 66), and the bloodmeal-induced increase in mosquito nitric oxide synthase gene expression was reduced by antibiotic treatment (11). Thus, AgISPR5 responded strongly to immune signals generated during bacterial infection and responded significantly but much less to parasite infection.
The mRNA for the putative regulatory protein AgIR28 was present at a very low level in untreated controls and was not detectably affected by antibiotic treatment (Fig. 4). Thus, the background level probably reflected constitutive rather than induced expression. AgIR28 mRNA was up-regulated more than 5-fold by LPS treatment as compared with the saline wounding control, and more than 100-fold over untreated mosquitoes. AgIR28 also responded markedly to bacteria in the normal bloodmeal. However, despite the strong up-regulation of AgIR28 mRNA by these bacterial elicitors, the gene displayed little additional response to parasite infection.
Figure 4.
Immune regulation of putative regulatory molecule AgIR28. Northern blot of poly(A)+ RNA isolated from 20 mosquitoes per lane after the indicated experimental treatment, probed with AgIR28. Mosquitoes were untreated (Control), 3-day antibiotic treatment (Antibiotic), 6 h after inoculation with sterile saline (Injury), 6 h after inoculation with LPS (LPS), 24 h after taking a normal uninfected bloodmeal (Normal), and 24 h after a P. berghei-infective bloodmeal (Infective). (Inset) Northern blot lanes in the same order probed with AgIR28 (Upper; 1.4 kb) and loading control rpS7 (Lower; 0.7 kb).
In distinction from the previous two genes, the gene for putative protease inhibitor AgIMcr14 displayed little response to bacterial infection (Fig. 5). Consistent with the low response by this gene to bacterial signals, bacterial proliferation in a normal bloodmeal also did not increase the mRNA level above that of untreated mosquitoes. Interestingly, however, AgIMcr14 mRNA abundance increased more than 8-fold during the period of ookinete invasion 24 h after an infective bloodmeal as compared with a normal bloodmeal. As an additional control, the same blot was reprobed with AgIR28 (Fig. 5 Inset). As in Fig. 4, AgIR28 again displayed a profile of expression that was essentially the inverse of AgIMcr14, namely much higher in response to bacterial infection as compared with parasite infection. The ratio between AgIR28 bacterial infection and wounding (3-fold at 6 h; Fig. 5 Inset, lower panel) was less pronounced than LPS inoculation versus sterile saline (5-fold at 6 h; Fig. 4). Nevertheless, it is clear that, overall, AgIMcr14 responded more strongly to parasite infection than did bacteria, and AgIR28, the reverse.
The mRNA levels of AgISPR5, AgIR28, and AgIMcr14 were equivalent after a normal bloodmeal or a bloodmeal infected with the gametocyte nonproducing 2.33 parasite strain (data not shown), indicating that the observed immune response was not a result of vertebrate host factors such as infection-related anemia or cytokines.
Taken together, these results indicate that molecular components of the mosquito innate immune system respond differentially to infection by bacteria and malaria parasites and that mosquito innate immunity can discriminate between signals generated by elicitors specific to bacterial or parasitic infections.
Discussion
In the current report, we present the results of an expression screen for genes up-regulated during the innate immune response of A. gambiae. LPS was used to stimulate immune signaling because it is a potent and general elicitor of innate immunity in insects and mammals (15, 67). It was also advantageous to use LPS rather than an actual pathogen for immune stimulation because sequences from the pathogen genome would have been coenriched along with up-regulated mosquito genes during solution hybridization. In addition, LPS produces a temporally discrete response, rather than a proliferative and persistent one like live bacteria, which should facilitate a temporal description of the mosquito immune response.
We do not know whether putative protease inhibitor AgIMcr14 expression was induced by recognition of parasite molecular patterns or by wounding during ookinete invasion of the midgut epithelium. In either case, the response was elevated after parasite infection as compared with a noninfective bloodmeal. Similarly, we do not know whether bacterial-response genes such as AgISPR5 were up-regulated during malaria infection because they recognized parasite molecular patterns or because the parasites carried bacteria as they penetrated midgut epithelial cells from the bloodmeal.
The major limitation of mRNA-based detection of molecular immune response is that only genes whose transcript abundance is altered will be detected. Posttranscriptional effects will be missed, as will genes that have no AluI or RsaI restriction fragments in the PCR-amplifiable size range. In the current study, we detected differences in the abundance of steady-state mRNA in response to LPS or pathogens, but further experiments would be necessary to determine whether the up-regulation resulted from new transcription or from mRNA stabilization.
It is likely that the insect innate immune response is a hierarchical gene expression program like the serum response of quiescent fibroblasts (43) and insect metamorphosis in response to ecdysone (44). The number of genes involved is not known, but it must be large. In the initial characterization of the enriched cDNA library reported here, only two genes were recovered more than once by random picking, and only two genes had been previously described. The enriched cDNA library is suitable for production of cDNA arrays for functional genomic analysis of the molecular circuitry of mosquito immunity.
Acknowledgments
We thank John Seed, Edge Biosystems, for helpful advice on properties of the binding matrices used. We acknowledge support to K.D.V. by the U. S. Public Health Service/National Institutes of Health (R01AI42361) and the United Nations Development Program/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases (970435).
Abbreviations
- LPS
bacterial lipopolysaccharide
- UTR
untranslated region
Note Added in Proof.
A recent report presented 19 A. gambiae genes that were inducible by bacterial infection (68). None of these genes were the same as any of the 23 immmune-inducible genes we report here. The strategies were different, however, because ref. 68 used LPS-stimulated hemocyte-like cultured cells, and we used LPS-stimulated whole mosquitoes. Thus, the cells used in ref. 68 represented a subset of the immune-responsive cells in whole mosquitoes and may not have included mRNAs expressed in other cell types, whereas cells related to those in ref. 68 and their cell-specific transcripts may be relatively rare in vivo and could therefore be rare in our enriched library made from whole-mosquito mRNA. However, the lack of overlap in the two collections also emphasizes the point that the insect immune response is complex and involves a large number of genes.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF283260–AF283275).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180060997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180060997
References
- 1.Warburg A, Miller L. Parasitol Today. 1991;7:179–181. doi: 10.1016/0169-4758(91)90127-a. [DOI] [PubMed] [Google Scholar]
- 2.Vernick K D. In: Molecular Mechanisms of Immune Response in Insects. Brey P T, Hultmark D, editors. London: Chapman & Hall; 1998. pp. 261–309. [Google Scholar]
- 3.Hogg J C, Hurd H. Parasitology. 1997;114:325–331. doi: 10.1017/s0031182096008542. [DOI] [PubMed] [Google Scholar]
- 4.Schiefer B A, Ward R A, Eldridge B F. Exp Parasitol. 1977;41:397–404. doi: 10.1016/0014-4894(77)90111-4. [DOI] [PubMed] [Google Scholar]
- 5.Hacker C S, Kilama W L. J Invertebr Pathol. 1974;23:101–105. doi: 10.1016/0022-2011(74)90079-2. [DOI] [PubMed] [Google Scholar]
- 6.Freier J E, Friedman S. J Invertebr Pathol. 1976;28:161–166. doi: 10.1016/0022-2011(76)90117-8. [DOI] [PubMed] [Google Scholar]
- 7.Klein T A, Harrison B A, Grove J S, Dixon S V, Andre R G. Bull W H O. 1986;64:901–907. [PMC free article] [PubMed] [Google Scholar]
- 8.Vaughan J A, Noden B H, Beier J C. Am J Trop Med Hyg. 1994;51:233–243. doi: 10.4269/ajtmh.1994.51.233. [DOI] [PubMed] [Google Scholar]
- 9.Beier J C. Annu Rev Entomol. 1998;43:519–543. doi: 10.1146/annurev.ento.43.1.519. [DOI] [PubMed] [Google Scholar]
- 10.Lowenberger C A, Kamal S, Chiles J, Paskewitz S, Bulet P, Hoffmann J A, Christensen B M. Exp Parasitol. 1999;91:59–69. doi: 10.1006/expr.1999.4350. [DOI] [PubMed] [Google Scholar]
- 11.Luckhart S, Vodovotz Y, Cui L, Rosenberg R. Proc Natl Acad Sci USA. 1998;95:5700–5705. doi: 10.1073/pnas.95.10.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins F H, Sakai R K, Vernick K D, Paskewitz S, Seeley D C, Miller L H, Collins W E, Campbell C C, Gwadz R W. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 13.Vernick K D, Fujioka H, Seeley D C, Tandler B, Aikawa M, Miller L H. Exp Parasitol. 1995;80:583–595. doi: 10.1006/expr.1995.1074. [DOI] [PubMed] [Google Scholar]
- 14.Gillespie J P, Kanost M R, Trenczek T. Annu Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 16.Dimopoulos G, Seeley D, Wolf A, Kafatos F C. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richman A M, Dimopoulos G, Seeley D, Kafatos F C. EMBO J. 1997;16:6114–6119. doi: 10.1093/emboj/16.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luckhart S, Rosenberg R. Gene. 1999;232:25–34. doi: 10.1016/s0378-1119(99)00121-3. [DOI] [PubMed] [Google Scholar]
- 19.Gorman M J, Andreeva O V, Paskewitz S M. Insect Biochem Mol Biol. 2000;30:35–46. doi: 10.1016/s0965-1748(99)00095-8. [DOI] [PubMed] [Google Scholar]
- 20.Lemaitre B, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaitre B, Nicolas E, Michaut L, Reichhart J M, Hoffmann J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 22.Benassi V, Coustau C, Carton Y. Arch Insect Biochem Physiol. 2000;43:64–71. doi: 10.1002/(SICI)1520-6327(200002)43:2<64::AID-ARCH2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 23.Dimopoulos G, Richman A, della Torre A, Kafatos F C, Louis C. Proc Natl Acad Sci USA. 1996;93:13066–13071. doi: 10.1073/pnas.93.23.13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton M G, Barker G C, Matsuoka H, Ramesar J, Janse C J, Waters A P, Sinden R E. Mol Biochem Parasitol. 1993;59:263–276. doi: 10.1016/0166-6851(93)90224-l. [DOI] [PubMed] [Google Scholar]
- 25.Kanamori A, Brown D D. Proc Natl Acad Sci USA. 1993;90:6013–6017. doi: 10.1073/pnas.90.13.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Brown D D. Proc Natl Acad Sci USA. 1991;88:11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duguid J R, Dinauer M C. Nucleic Acids Res. 1990;18:2789–2792. doi: 10.1093/nar/18.9.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forster A C, McInnes J L, Skingle D C, Symons R H. Nucleic Acids Res. 1985;13:745–761. doi: 10.1093/nar/13.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franz O, Bruchhaus I, Roeder T. Nucleic Acids Res. 1999;27:e3. doi: 10.1093/nar/27.11.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zibara K, Bourdillon M C, Chignier E, Covacho C, McGregor J L. Arterioscler Thromb Vasc Biol. 1999;19:1650–1657. doi: 10.1161/01.atv.19.7.1650. [DOI] [PubMed] [Google Scholar]
- 31.Hung H L, Song F, Gewirtz A. Leukemia. 1999;13:295–297. doi: 10.1038/sj.leu.2401274. [DOI] [PubMed] [Google Scholar]
- 32.Guiguen Y, Baroiller J F, Ricordel M J, Iseki K, McMeel O M, Martin S A, Fostier A. Mol Reprod Dev. 1999;54:154–162. doi: 10.1002/(SICI)1098-2795(199910)54:2<154::AID-MRD7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Richman A M, Bulet P, Hetru C, Barillas-Mury C, Hoffmann J A, Kafalos F C. Insect Mol Biol. 1996;5:203–210. doi: 10.1111/j.1365-2583.1996.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 34.Salazar C E, Mills-Hamm D, Kumar V, Collins F H. Nucleic Acids Res. 1993;21:4147. doi: 10.1093/nar/21.17.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Salazar C E, Hamm D M, Wesson D M, Beard C B, Kumar V, Collins F H. Insect Mol Biol. 1994;3:1–13. doi: 10.1111/j.1365-2583.1994.tb00145.x. [DOI] [PubMed] [Google Scholar]
- 37.Kanost M R, Jiang H. Adv Exp Med Biol. 1997;425:155–161. doi: 10.1007/978-1-4615-5391-5_15. [DOI] [PubMed] [Google Scholar]
- 38.Potempa J, Korzus E, Travis J. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 39.Levashina E A, Langley E, Green C, Gubb D, Ashburner M, Hoffmann J A, Reichhart J M. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- 40.Kanost M R. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 41.Iwaki D, Kawabata S, Miura Y, Kato A, Armstrong P B, Quigley J P, Nielsen K L, Dolmer K, Sottrup-Jensen L, Iwanaga S. Eur J Biochem. 1996;242:822–831. doi: 10.1111/j.1432-1033.1996.0822r.x. [DOI] [PubMed] [Google Scholar]
- 42.Armstrong P B, Quigley J P. Dev Comp Immunol. 1999;23:375–390. doi: 10.1016/s0145-305x(99)00018-x. [DOI] [PubMed] [Google Scholar]
- 43.Lau L F, Nathans D. EMBO J. 1985;4:3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fletcher J C, Thummel C S. Development (Cambridge, UK) 1995;121:1411–1421. doi: 10.1242/dev.121.5.1411. [DOI] [PubMed] [Google Scholar]
- 45.Valenzuela J G, Francischetti I M, Ribeiro J M. Biochemistry. 1999;38:11209–11215. doi: 10.1021/bi990761i. [DOI] [PubMed] [Google Scholar]
- 46.Arca B, Lombardo F, de Lara Capurro M, della Torre A, Dimopoulos G, James A A, Coluzzi M. Proc Natl Acad Sci USA. 1999;96:1516–1521. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brightbill H D, Libraty D H, Krutzik S R, Yang R B, Belisle J T, Bleharski J R, Maitland M, Norgard M V, Plevy S E, Smale S T, Brennan P J, Bloom B R, Godowski P J, Modlin R L. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 48.Wittwer D, Wiesner A. Arch Insect Biochem Physiol. 1998;39:91–97. doi: 10.1002/(SICI)1520-6327(1998)39:3<91::AID-ARCH1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 49.Ashida M, Brey P. In: Molecular Mechanisms of Immune Response in Insects. Brey P T, Hultmark D, editors. London: Chapman & Hall; 1998. pp. 135–172. [Google Scholar]
- 50.Jiang H, Kanost M R. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 51.Kawabata S, Tokunaga F, Kugi Y, Motoyama S, Miura Y, Hirata M, Iwanaga S. FEBS Lett. 1996;398:146–150. doi: 10.1016/s0014-5793(96)01224-0. [DOI] [PubMed] [Google Scholar]
- 52.Dimopoulos G, Richman A, Muller H M, Kafatos F C. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura A, Kawasaki K, Kubo T, Natori S. Int J Dev Biol. 1992;36:391–398. [PubMed] [Google Scholar]
- 54.Kitabayashi A N, Arai T, Kubo T, Natori S. Insect Biochem Mol Biol. 1998;28:785–790. doi: 10.1016/s0965-1748(98)00058-7. [DOI] [PubMed] [Google Scholar]
- 55.Hart C M, Laemmli U K. Curr Opin Genet Dev. 1998;8:519–525. doi: 10.1016/s0959-437x(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 56.Nappi A J, Vass E, Frey F, Carton Y. Eur J Cell Biol. 1995;68:450–456. [PubMed] [Google Scholar]
- 57.Nappi A J, Vass E. Pigment Cell Res. 1993;6:117–126. doi: 10.1111/j.1600-0749.1993.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 58.Hufton S E, Jennings I G, Cotton R G. Biochem J. 1995;311:353–366. doi: 10.1042/bj3110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Zhao X, Christensen B M. Insect Biochem Mol Biol. 1994;24:1043–1049. doi: 10.1016/0965-1748(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 60.Heine H, Delude R L, Monks B G, Espevik T, Golenbock D T. J Biol Chem. 1999;274:21049–21055. doi: 10.1074/jbc.274.30.21049. [DOI] [PubMed] [Google Scholar]
- 61.Morimoto R I. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 62.Mehlen P, Kretz-Remy C, Preville X, Arrigo A P. EMBO J. 1996;15:2695–2706. [PMC free article] [PubMed] [Google Scholar]
- 63.Arrigo A P. Biol Chem. 1998;379:19–26. [PubMed] [Google Scholar]
- 64.Putsep K, Branden C I, Boman H G, Normark S. Nature (London) 1999;398:671–672. doi: 10.1038/19439. [DOI] [PubMed] [Google Scholar]
- 65.Putsep K, Normark S, Boman H G. FEBS Lett. 1999;451:249–252. doi: 10.1016/s0014-5793(99)00582-7. [DOI] [PubMed] [Google Scholar]
- 66.Pumpuni C B, Beier M S, Nataro J P, Guers L D, Davis J R. Exp Parasitol. 1993;77:195–199. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- 67.Hoffmann J A, Reichart J-M. Trends Cell Biol. 1997;7:309–316. [Google Scholar]
- 68.Dimopoulos G, Casavant T L, Chang S R, Scheetz T, Roberts C, Donohue M, Schultz J, Benes V, Bork P, Ansorge W, et al. Proc Natl Acad Sci USA. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]