Abstract
The Tol2 element of the medaka fish Oryzias latipes belongs to the hAT family of transposons (hobo/Ac/Tam3). We report here identification of a functional transposase of Tol2 that is capable of catalyzing its transposition in the germ line of zebrafish Danio rerio. A transcript produced from Tol2 encodes a putative transposase. Zebrafish fertilized eggs were coinjected with mRNA transcribed in vitro, using cDNA of the Tol2 transcript as a template and a plasmid DNA harboring a mutant Tol2, which had a deletion in the putative transposase gene but retained necessary cis sequences. The injected fish were raised to adulthood and mated to noninjected fish, and genomic DNA of the progeny fish were analyzed by PCR and Southern hybridization. Half of F1 fish obtained from one of eight injected fish contained the Tol2 DNA in their genomes but not the vector portion. Among these F1 fish, Tol2 insertions at four different loci were identified, and some F1 fish carried two or three different Tol2 insertions, indicating that the germ line of the founder fish is highly mosaic. Sequencing analyses revealed that, in all cases, Tol2 was surrounded by zebrafish genomic sequences, and an 8-bp duplication was created at the target site, indicating that Tol2 was integrated in the zebrafish genome through transposition. This study identifies an autonomous member of a DNA-based transposable element from a vertebrate genome. The Tol2 transposon system should thus be used to develop novel transgenesis and insertional mutagenesis methods in zebrafish and possibly in other fishes.
The DNA-based transposable elements (or transposons) are repetitive sequences that move from one locus in the genome to another and have been used as powerful tools to study model animals and plants. The members of transposable elements fall into two classes: 1) autonomous elements, such as the Ac element in maize, encode a functional transposase and can transpose by itself; 2) nonautonomous elements, such as the Ds element in maize, carry mutations or deletions in the transposase gene and can be mobilized only in the presence of autonomous members (1–7). In vertebrates, several repetitive sequences similar to transposons of the Tc1/mariner elements of Caenorhabditis elegans and Drosophila have been identified (8–12). Although a synthetic Sleeping Beauty transposase, which was reconstructed based on a consensus sequence derived from sequences of nonautonomous salmonid Tc-like elements, has been shown to be capable of catalyzing transposition in vertebrate cells (13), an endogenous autonomous element has not yet been identified from a vertebrate genome.
The Japanese medaka fish, Oryzias latipes, is a freshwater teleost inhabiting East Asia. More than 80 mutations that affect pigmentation and body shapes have been isolated mainly from wild populations in Japan (ref. 14; see also http://biol1.bio.nagoya-u.ac.jp:8000/). Among them, mutations in the i locus, which encodes a gene for tyrosinase, cause amelanotic skin and red-colored eyes (15). In one of the i alleles, i4, an insertion of about 4.7 kilobases (kb) of DNA was found in the fifth exon of the tyrosinase gene. The sequence of the insertion is similar to those of transposons of the hAT family; i.e., hobo of Drosophila (16, 17), Ac of maize (4, 5), and Tam3 of snapdragon (18, 19). The element was named Tol2 (20). Laboratory strains of the medaka fish contain about ten copies of the Tol2 element per haploid genome. The Tol2-tyr element, a particular Tol2 element which resides in the tyrosinase gene, has been shown by PCR to be excisable from the target locus during embryogenesis, suggesting the presence of at least one autonomous member somewhere in the genome (20). Until now, neither a functional transposase nor an autonomous member of the Tol2 element, however, had been identified.
Zebrafish, Danio rerio, is another small teleost that has been used as a model animal to study vertebrate genetics and development. The zebrafish genome does not contain the Tol2 element (21). To determine whether the Tol2 element encodes a functional transposase, we have been investigating the activities of the Tol2-tyr element using zebrafish (21, 22). In our previous studies, we cloned cDNAs of the Tol2-tyr element from zebrafish embryos injected with a plasmid DNA harboring the Tol2-tyr element and developed a transient embryonic excision assay. In this assay, zebrafish fertilized eggs were coinjected with mRNA transcribed in vitro using the full-length cDNA as a template and a plasmid DNA harboring a mutant Tol2 element, which has a deletion in the putative transposase coding region; DNA extracted from the injected embryos at 50–100% epiboly stages was analyzed by PCR. The mutant nonautonomous Tol2 element is excisable from the vector plasmid only when coinjected with the mRNA, indicating that the mRNA can produce a putative transposase activity that catalyzes the excision reaction.
In the present study, we aimed to determine whether the Tol2-tyr element encodes a fully functional transposase that catalyzes a complete transposition process, excision and reintegration, in zebrafish. We show here that mRNA transcribed from the Tol2-tyr element produces a fully functional transposase, which is capable of catalyzing transposition of a nonautonomous Tol2 element in the zebrafish germ lineage. The Tol2-tyr element is thus shown to encode a functional transposase, and, hence, to be autonomous. We also suggest that the Tol2 transposon system should be useful to develop transgenesis and insertional mutagenesis methods in zebrafish and possibly in other fish species.
Materials and Methods
Fish.
Zebrafish strains, Tübingen, TL (leot1, lofdt2) and brass, were used for microinjection and mating.
Plasmids and Primers.
The structures of the (Tol2-tyr)ΔRV plasmid (21) and the Tol2 cDNA (22) are shown in Fig. 1. The cDNA of 2149 bp was cloned into the XhoI site of pBlueScript II SK+ (Stratagene) and used for in vitro transcription. The positions and directions of primers used in this study are shown in Fig. 1 and Fig. 3 A and C. Nucleotide sequences of tyr-ex5r, tyr-ex4f, and the zebrafish wnt5A primers were described previously (21–24). The sequences of other primers are as follows. Tol2f1, 5′-CUACUACUACUAAGACATTCCGCTGCACTTGCC-3′;. Tol2f2, 5′-CUACUACUACUAACTTGTACTTTCACTTGAGTA-3′; Tol2f5, 5′-CTGCTCTGATCATGAAACAG-3′; Tol2f20, 5′-TTTACTCAAGTAAGATTCTAG-3′; Tol2r1, 5′-CUACUACUACUACAAGTAAAGTAAAAATCCCCA-3′; Tol2r2, 5′-CUACUACUACUAGCAAAGAAAGAAAACTAGAGA-3′; Tol2r7, 5′-CCGATGCGGGAAGAGGTGTATTAG-3′; Tol2r9, 5′-GGAAAATAGAATGAAGTGATCTCC-3′; Tol2r10, 5′-CTCCATTAAAATTGTACTTGA-3′; 7a5′, 5′-GACGTAGTGTCCTGCTGGTGTA-3′; 7a3′, 5′-CGAGAAGCAGCGCCTGGAGCGA-3′; 7b5′, 5′-GTTTTATGCAAAAGTTTGGCC-3′; 7b3′, 5′-TCTCGGCCTGTTGCAGCAGAG-3′; 7c5′, 5′-CAATATTGTGCTGAAGGCTACA-3′; 7c3′, 5′-CTGGTTAGTCAATGCCATAGT-3′; 7d5′, 5′-CAAATCATTGTGCATGCATGTA-3′; 7d5′-2, 5′-AGAGCAAATTTGTTCTTCTGG-3′; and 7d3′, 5′-CCTTCCCCTTCACACACTTGGTT-3′.
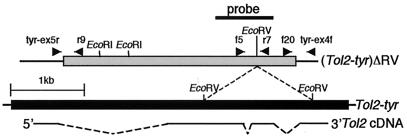
Figure 1.
Structures of the Tol2 elements and cDNA. The black and gray boxes indicate the Tol2-tyr and (Tol2-tyr)ΔRV elements, respectively, and thin lines flanking these boxes represent plasmid DNA (the medaka fish tyrosinase sequence). Self-ligation at two EcoRV sites in Tol2-tyr generated (Tol2-tyr)ΔRV. 5′ and 3′ in Tol2 cDNA indicate the direction of transcription, and dotted lines indicate introns. Arrowheads show positions and directions of primers used in the experiment described in Fig. 2A. A thick bar shows the probe used in Southern hybridization analyses. Note that the total length of the Tol2-tyr element, previously reported as 4681 bp (20), has been corrected to 4682 bp (D84375), by addition of 1 bp in the first intron, by the authors of the first report. r9, Tol2r9; f5, Tol2f5; r7, Tol2r7; f20, Tol2f20.
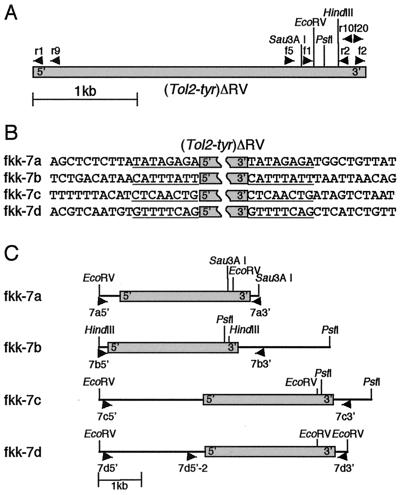
Figure 3.
Inverse PCR and DNA sequencing analysis of the Tol2 insertions. (A) Primers and restriction enzyme sites used for inverse PCR. The gray box indicates (Tol2-tyr)ΔRV. Arrowheads indicate positions and directions of primers. EcoRV and HindIII are unique sites. Sau3A I and PstI cut more than twice, and only most 3′ sites used to clone the 3′ junctions are drawn. r1, Tol2r1; r9, Tol2r9; f5, Tol2f5; f1, Tol2f1; r2, Tol2r2; r10, Tol2r10; f20, Tolf20; f2, Tol2f. (B) DNA sequences adjacent to (Tol2-tyr)ΔRV. DNA fragments were amplified by inverse PCR from fish with fkk-7a, fkk-7b, fkk-7c, and fkk-7d insertions (Fig. 2C, lanes 1–4), and sequenced. Eight-base pair duplications, created upon transposition, are underlined. (C) Structures of Tol2 insertions. These structures were drawn based on DNA sequences from inverse PCR products and the wild-type DNA fragments. The gray box indicates (Tol2-tyr)ΔRV. Restriction enzyme sites used for inverse PCR are shown. Arrowheads (7a5′ and 7a3′, 7b5′ and 7b3′, 7c5′ and 7c3′, and 7d5′ and 7d3′) indicate positions and directions of primers used to amplify wild-type DNA fragments, which extend across the integration sites for Tol2. 7d5′-2 primer was used in the experiment described in Fig. 4.
In Vitro Transcription and Microinjection.
In vitro transcription of Tol2 mRNA using T7 RNA polymerase and microinjection of the mRNA and the (Tol2-tyr)ΔRV plasmid DNA were carried out as described previously (22).
Southern Hybridization and PCR Analyses of F1 Fish.
For PCR analysis of pooled F1 embryos, 20–30 embryos of 1 day old were pooled in one microtube and lysed in 300 μl of DNA extraction buffer (10 mM EDTA/10 mM Tris⋅HCl, pH 8.0/200 μg/ml proteinase K) at 50°C for 3 h or overnight. Genomic DNA was extracted with phenol/chloroform, precipitated with ethanol, and resuspended in 50 μl of 10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA. An aliquot of 1 μl was used for PCR: 35 cycles of 20 s, 94°C; 20 s, 56°C; 20 s, 72°C with Tol2f5 and Tol2r7. For PCR analysis of adult F1 fish, caudal fins were clipped and lysed in 200 μl of DNA extraction buffer at 50°C for 3 h or overnight. Genomic DNA was extracted with phenol/chloroform, precipitated with ethanol, and resuspended in 50 μl of 10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA. An aliquot of 1 μl was used for PCR: 30 cycles of 20 s, 94°C; 20 s, 56°C; 20 s, 72°C with primers described in the text. For Southern hybridization, 5 μg of caudal fin DNA of F1 fish was digested either with EcoRV or EcoRI, separated on a 1% agarose gel, transferred on a nylon membrane, and hybridized with the 32P-labeled probe shown in Fig. 1.
Inverse PCR and DNA Sequencing.
One microgram of caudal fin DNA isolated from F1 fish was digested with EcoRV, PstI, HindIII, or Sau3A I, diluted to 2 ng/ml, and then circularized by ligation. The ligated DNA was concentrated by ethanol precipitation and used for two rounds of PCR; i.e., first PCR: 30 cycles of 30 s, 94°C; 30 s, 58°C; 5 min, 72°C with Tol2f5 and Tol2r9 (for 5′-end) or with Tol2f20 and Tol2r10 (for 3′-end); an aliquot was used for the second PCR: 30 cycles of 30 s, 94°C; 30 s, 58°C; 5 min, 72°C with Tol2f1 and Tol2r1 (for 5′-end) or with Tol2f2 and Tol2r2 (for 3′-end). The amplified DNA were gel-extracted and cloned using pAmp10 system (GIBCO/BRL). For cloning of wild-type DNA, 100 ng of genomic DNA was used for PCR: 30 cycles of 30 s, 94°C; 30 s, 56°C; 5 min, 72°C with primers shown in Fig. 3C. The amplified DNA were gel-extracted and cloned using TOPO TA Cloning (Invitrogen). The plasmid DNA was prepared from bacteria and sequenced using BigDye terminator cycle sequencing kit (Applied Biosystems) and ABI PRISM 310 genetic analyzer.
Results
Identification of Founder Fish.
A transcript of 2156 nt is synthesized from the Tol2-tyr element in zebrafish embryos injected with the Tol2-tyr plasmid DNA (Fig. 1) (22). The cDNA of 2149 bp was inserted into a pBlueScript II plasmid, which lacked the first 7 bp of the 5′-end (5′-ACGTCATGTC … ). This eliminates the possibility for the first ATG, located at nucleotides 6–8, to be used as a start codon, which would produce a short peptide of 11 aa. The resulting cDNA has the capacity to encode a protein of 649 aa when the first ATG was used as a start codon. The (Tol2-tyr)ΔRV element has a deletion of ≈1.5 kb, which inactivates the putative transposase by disrupting the exon sequences and hence becomes a nonautonomous element (Fig. 1) (21, 22).
To test whether the Tol2 transcript encodes a functional transposase that catalyzes transposition in the zebrafish germ lineage, zebrafish fertilized eggs were coinjected with mRNA transcribed in vitro using the plasmid DNA carrying the Tol2 cDNA as a template and the (Tol2-tyr)ΔRV plasmid DNA. It is expected that a transposase is synthesized from the mRNA, excises the (Tol2-tyr)ΔRV element from the plasmid DNA, and reintegrates it into the genome of the zebrafish germ cells during embryogenesis. Such transposition events could be detected by analyzing F1 progeny fish from the injected fish. In every injection experiment, aliquots of the injected embryos were subjected to a transient excision assay (22), and the excision activity was verified (data not shown). The rest of the injected fish were raised to adulthood, and pair mating to noninjected fish of opposite sexes yielded F1 offspring. Eight injected fish mated successfully, and at least 50 embryos from each mating were pooled and analyzed by PCR using the Tol2f5 and Tol2r7 primers for the presence of the (Tol2-tyr)ΔRV DNA. Pools of F1 embryos from two of eight fish were PCR positive (data not shown). Therefore, the F1 progeny fish from these two founder fish, referred to as fkk-1 and fkk-7, were raised to adulthood and studied further.
Analysis of F1 Fish with Insertions of Tol2.
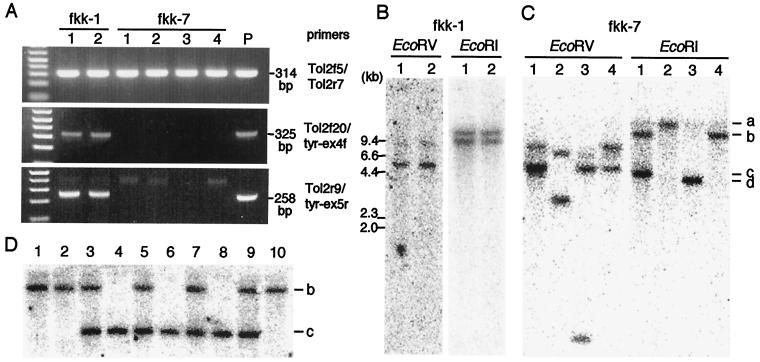
Genomic DNA isolated from a caudal fin of individual F1 fish was analyzed by PCR using primer pairs Tol2f5 and Tol2r7, Tol2f20 and tyr-ex4f, and Tol2r9 and tyr-ex5r (Figs. 1 and 2A). Two fish out of 68 F1 fish from the fkk-1 founder fish were positive for the Tol2 sequence (fkk-1, lanes 1 and 2 of Tol2f5/Tol2r7, Fig. 2A), and these fish were shown to carry the tyrosinase gene sequences of the medaka fish flanking the Tol2 sequence (fkk-1, lanes 1 and 2 of Tol2f20/tyr-ex4f and Tol2r9/tyr-ex5r, Fig. 2A). In contrast, 25 fish out of 50 F1 fish from the fkk-7 founder fish, which were positive for the Tol2 sequence, did not carry the tyrosinase gene sequences flanking Tol2 (four representatives of the PCR analyses are shown in fkk-7, lanes 1–4 of Tol2f5/Tol2r7, Tol2f20/tyr-ex4f, and Tol2r9/tyr-ex5r, Fig. 2A). The (Tol2-tyr)ΔRV DNA injected into fertilized eggs could integrate into the genome either through nonhomologous recombination, independently of a transposase activity (25), or, as expected, in a transposase-dependent manner. In the former case, the Tol2 sequence is likely to be flanked by sequences of the vector plasmid, the tyrosinase gene sequences of the medaka fish; in the latter case, it should be surrounded by zebrafish genomic sequences. Thus, in F1 fish from fkk-1, the integration was likely to occur independently of transposition.
Figure 2.
PCR and Southern hybridization analyses of F1 fish. (A) PCR analyses of genomic DNA extracted from caudal fin clippings of F1 fish from fkk-1 and fkk-7. Primer pairs used for PCR and the sizes of the PCR products are shown on the right. Two (fkk-1, lanes 1 and 2) and 25 F1 fish were found positive for PCR using Tol2f5/Tol2r7 out of 68 and 50 F1 fish from fkk-1 and fkk-7, respectively. PCR analyses of the 25 F1 fish from fkk-7 yielded the same results, and four representatives are shown (fkk-7, lanes 1–4). The same four samples were used also for Southern hybridization (described in C). In the control (P), 100 ng of zebrafish genomic DNA plus 1 pg of the (Tol2-tyr)ΔRV plasmid DNA were used as a PCR template. (B) Southern hybridization analysis of F1 fish from fkk-1. DNA samples of two fish (A) were digested with either EcoRV or EcoRI and hybridized with the probe shown in Fig. 1. (C) Southern hybridization analysis of F1 fish from fkk-7. DNA samples of four fish (A) were digested with either EcoRV or EcoRI and hybridized with the probe (Fig. 1). EcoRI bands of four different sizes, shown as a–d on the right, correspond to the fkk-7a, fkk-7b, fkk-7c, and fkk-7d insertions described in the text. The lower bands in EcoRV lane 1 are probably a triplet. (D) Southern hybridization analysis of F2 fish from the lane 1 F1 fish (fkk-7). Ten fish out of 14 F2 fish, which were positive for PCR using Tol2f5 and Tol2r7, were analyzed. DNA samples were digested with EcoRI and hybridized with the probe (Fig. 1). Fish with fkk-7b alone (lanes 1, 2, 10), fkk-7b alone (lanes 4, 6, 8), and both fkk-7b and fkk-7c (lanes 3, 5, 7, 9) were identified.
Digestion of the genomic DNA containing the (Tol2-tyr)ΔRV DNA with EcoRV and EcoRI should give rise to two bands and a single band in Southern hybridization using the probe shown in Fig. 1, respectively. Southern hybridization analysis of the genomic DNA of the two transgenic F1 fish from fkk-1 showed the same hybridization pattern either by digestion with EcoRV or EcoRI (Fig. 2B). Although these fish might have the Tol2 DNA at two distinct loci and one of them might integrate through transposition, these possibilities were not examined further. On the other hand, Southern hybridization analyses of the F1 fish from fkk-7 showed DNA bands of four different sizes by digestion with EcoRI (Fig. 2C), indicating that at least four different integration events of the Tol2 DNA took place in the germ line of fkk-7. These Tol2 insertions were designated as fkk-7a, fkk-7b, fkk-7c, and fkk-7d with respect to the sizes of the EcoRI bands (Fig. 2C). Although the F1 fish on lanes 2, 3, and 4 carried a single Tol2 insertion, fkk-7a, fkk-7d, and fkk-7b, respectively, the F1 fish on lane 1 was likely to carry two different Tol2 insertions (Fig. 2C, lane 1). To verify this observation, the lane 1 F1 fish was crossed with wild-type fish, and F2 offspring were raised. Ten fish out of 14 F2 fish were positive for PCR using Tol2f5 and Tol2r7 (data not shown), and these 10 fish were then analyzed by Southern hybridization (Fig. 2D, lanes 1–10). The result indicates that the lane 1 F1 fish carried both the fkk-7b and fkk-7c insertions, which were generated at distinct loci in the zebrafish genome.
Evidences for Transposition of Tol2.
The structures of the insertions identified among the F1 fish from fkk-7 were analyzed by inverse PCR (Fig. 3A). For fkk-7a, DNA fragments containing the 5′- and 3′-ends (with respect to the direction of transcription) of the (Tol2-tyr)ΔRV element were amplified from genomic DNA of the lane 2 fish (Fig. 2C) digested with EcoRV and Sau3A I, respectively, and sequenced (Fig. 3B, fkk-7a). For fkk-7b, DNA fragments containing the 5′- and 3′-ends of the Tol2 element were amplified from genomic DNA of the lane 4 fish (Fig. 2C) digested with HindIII and PstI, respectively, and sequenced (Fig. 3B, fkk-7b). For fkk-7c, DNA fragments containing the 5′- and 3′-ends of the Tol2 element were amplified from genomic DNA of the lane 1 fish (Fig. 2C) digested with EcoRV and PstI, respectively, and sequenced (Fig. 3B, fkk-7c). Although this fish carries both fkk-7b and fkk-7c, the sequences of the DNA fragments amplified by inverse PCR using those restriction enzymes are distinct from those obtained for the fkk-7b insertion and hence are thought to correspond to fkk-7c. For fkk-7d, DNA fragments containing the 5′- and 3′-ends of the Tol2 element were amplified from genomic DNA of the lane 3 fish (Fig. 2C) digested with EcoRV and sequenced (Fig. 3B, fkk-7d). All of these 5′ and 3′ sequences contained the same 8-bp sequences adjacent to the Tol2 sequence. It has been shown that an 8-bp duplication is created at the target site upon transposition of transposons of the hAT family (4, 5, 16, 18, 20). Thus, these results strongly suggest that, in all four cases, the integration of the Tol2 element was carried out through transposition.
To confirm this notion, we characterized wild-type alleles of these target loci. We constructed primer pairs 7a5′ and 7a3′, 7b5′ and 7b3′, 7c5′ and 7c3′, and 7d5′ and 7d3′ (Fig. 3C), based on the DNA sequences obtained from the inverse PCR analyses, so as to amplify DNA fragments extending across the integration sites of the Tol2 element. PCR using these primer pairs and genomic DNA isolated from a wild-type zebrafish strain successfully amplified DNA fragments. Sequencing analyses of these DNA fragments revealed that each wild-type locus contained a single 8-bp sequence (wild-type sequences for fkk-7a, fkk-7b, fkk-7c, and fkk-7d are AB045573, AB045574, AB045575, and AB045576, respectively), which was duplicated in the F1 fish, and the integration of the Tol2 element did not cause any chromosomal rearrangement at the target locus. Therefore, we conclude that these Tol2 insertions were generated through transposition and that the mRNA injected with the (Tol2-tyr)ΔRV DNA can produce a fully functional transposase, which catalyzes a complete transposition process, excision and reintegration reactions, in the zebrafish germ lineage. The structures of the Tol2 insertions are summarized in Fig. 3C. The ≈3-kb, ≈5-kb, and ≈5-kb and ≈0.8-kb EcoRV fragments in fkk-7a, fkk-7c, and fkk-7d shown in Fig. 3C are consistent with the sizes of the DNA bands observed in the Southern hybridization analysis (Fig. 2C, EcoRV lanes 1–3).
Multiple Tol2 Insertions in F1 Fish.
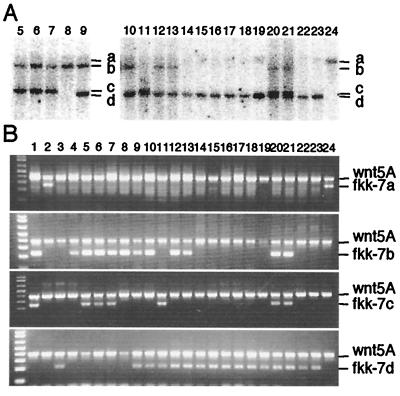
Genomic DNA of 24 F1 fish (25 fish were originally identified as PCR positives as described above, but one fish was lost before this analysis) from fkk-7 were analyzed by Southern hybridization (Fig. 2C, lanes 1–4, and Fig. 4A, lanes 5–24). The individual F1 fish had one (Fig. 2C, lanes 2–4, and Fig. 4A, lanes 8, 14–19, 22–24), two (Fig. 2C, lane 1, and Fig. 4A, lanes 5–7, 9–13), or three (Fig. 4A, lanes 20 and 21) insertions, which should correspond to any of the insertions identified above. To confirm the identities of the insertions, PCR analyses were performed using primers designed to detect specific Tol2 insertions; i.e., for fkk-7a, 7a3′ and Tol2f20; for fkk-7b, 7b5′ and Tol2r1; for fkk-7c, 7c3′ and Tol2f20; for fkk-7d, 7d5′-2 and Tol2r1 (Fig. 4B). The results are also summarized in Table 1. Thus, the four Tol2 insertions were segregated independently in the germ line of the fkk-7 founder fish, giving rise to the highly mosaic germ line, and we could obtain F1 fish with multiple insertions, up to three insertions.
Figure 4.
Identification of the Tol2 insertions in F1 fish. (A) Southern hybridization analysis of F1 fish from fkk-7. DNA samples from 20 F1 fish were digested with EcoRI and hybridized with the probe (Fig. 1). The numbering of the lanes (fish) is continued from Fig. 2C. Bands of four different sizes, a (lane 24), b (lanes 5–10, 12, 13, 20, 21), c (lanes 5–7, 11, 20, 21), and d (lanes 9–23), were detected. The lower bands on lanes 11, 20, and 21 are doublets. (B) PCR analysis of 24 F1 fish from fkk-7. DNA samples on lanes 1–4 correspond to those on lanes 1–4 of Fig. 2C. PCR reactions were carried out using the wnt5A primers (positive control) and 7a3′ and Tol2f20 (fkk-7a), 7b5′ and Tol2r1 (fkk-7b), 7c3′ and Tol2f20 (fkk-7c), or 7d5′-2 and Tol2r1 (fkk-7d). On these photos, the presence of the lower band indicates that the fish carries the specific Tol2 insertion. The results are consistent with those obtained by Southern hybridization analysis and are summarized in Table 1.
Table 1.
Classification of transgenic F1 fish
| Founder fish | Transgenic fish*/F1 progeny fish | Type of integration | Grouping by Southern hybridization and PCR | |
|---|---|---|---|---|
| fkk-1 | 2/68 | Illegitimate† | Same | 2/2 |
| fkk-7 | 25/50 | Transposition | a alone | 2/24‡ |
| b alone | 2/24 | |||
| d alone | 9/24 | |||
| b + c | 4/24 | |||
| b + d | 4/24 | |||
| c + d | 1/24 | |||
| b + c + d | 2/24 | |||
Transgenic fish were identified as positives for PCR using Tol2f5 and Tol2r7.
The structure of the insertions in F1 fish from fkk-1 has not been analyzed by DNA cloning.
Discussion
In the present study, we have demonstrated that a nonautonomous Tol2 element could transpose in the genome of the zebrafish germ lineage when coinjected with mRNA transcribed from the Tol2-tyr element. Therefore, we concluded that the Tol2-tyr element is an autonomous member that encodes a functional transposase capable of catalyzing transposition. In vertebrates, several repetitive sequences similar to transposons of the Tc1/mariner family have been reported (8–12). Although a synthetic Sleeping Beauty transposase, which was reconstructed based on a consensus sequence obtained from nonautonomous salmonid Tc-like elements, has been shown to be capable of catalyzing transposition in vertebrate-cultured cells (13), neither an autonomous member nor a functional transposase has yet been identified from endogenous transposable elements in vertebrates. This study identified an autonomous member of a DNA transposable element from a vertebrate genome. Also, we have demonstrated that controlled germ-line transformation in zebrafish can be mediated by using the Tol2 transposable element.
We have previously shown by a transient embryonic excision assay that the (Tol2-tyr)ΔRV element is excisable but the Tol2 element lacking the first intron sequence of the transposase gene cannot be excised, and we have suggested the presence of essential cis elements in this region (22). It has, however, not been clear whether the (Tol2-tyr)ΔRV sequence is sufficient for the reintegration reaction. Our present result clearly indicates that the (Tol2-tyr)ΔRV element contains cis elements necessary for transposition. Further studies on dissecting and defining cis requirements will be needed to develop useful transposon vectors. Host factors necessary for the transposition reaction of the Tol2 element other than the transposase itself (e.g., polymerase, exonuclease, ligase, etc.) have not been known. Our study suggests that such host factors are conservatively present in both zebrafish and the medaka fish, which diverged from a common ancestor some 150 million years ago (26). Therefore, the Tol2 transposon system may possibly work also in other fish species.
The members of the hAT family are known to comprise complete and defective copies in the genome, which correspond to autonomous and nonautonomous elements, respectively. The defective copies are often shorter, as they have deletions of different sizes in the transposase coding region (3–7). This is not the case with Tol2 because it has been shown that members of the Tol2 element in the medaka fish genome are highly homogeneous in their structures, and some of them are identical to the Tol2-tyr element at the sequence level (27). These observations, together with our present results, raise a question whether all, or most, of the Tol2 element in the medaka fish genome encodes a functional transposase. It has been shown that the excision frequency of an endogenous Tol2 element is relatively low (20). In this regard, it should be noted that Koga et al. isolated mRNA from the medaka fish cells and identified a longer (2319 nt) and a shorter (1946 nt) transcript transcribed from anonymous Tol2 elements (28). Given the first ATG was used for translation initiation, the longer transcript has the capacity to encode a protein of 685 aa, and the shorter one encodes a protein of 576 aa. These are different from the protein of 649 aa, which could be synthesized from mRNA used in this study and was shown to be fully functional. The activities of the mRNAs transcribed endogenously in the medaka fish cells have not yet been reported. Studies on those activities using the zebrafish system described here should give an insight into the regulation of the Tol2 element in the medaka fish.
Construction of transgenic animals is an important technology to study functions of genes and also to develop insertional mutagenesis methods. Germ-line transmission of foreign DNA has been achieved in zebrafish by microinjection of naked DNA into fertilized eggs (25), by infection of a pseudotyped retrovirus to blastula-stage embryos (23), and by using transposon systems derived from Tc3 of C. elegans (29) and mariner of Drosophila (30), both members of the Tc1/mariner family. Microinjection of DNA is the most popular way to construct transgenic zebrafish and has been applied to study the function of promoters in vivo (31, 32). The pseudotyped retrovirus system can generate a large number of insertions at different loci very efficiently (33) and has made it possible for a large-scale insertional mutagenesis to be performed (34–38). In contrast to these, development of transposon technologies in zebrafish is still in its early stages, and neither highly efficient transgenesis nor insertional mutagenesis using a transposon has been established yet. In the case of Tc3, the Tc3 vector DNA and mRNA for the Tc3 transposase were coinjected into fertilized eggs, and only one transposase-mediated integration event was identified among F1 fish from 40 injected fish (29). In the case of mariner, the mariner (peach) vector DNA and mRNA for the mariner transposase were coinjected into fertilized eggs, and four transposase-mediated integration events were identified among F1 fish from 12 injected fish by Southern hybridization analysis, but not at the DNA sequence level (30). Our present study clearly identified four different transposition events among F1 fish from eight injected fish. Although the relatively small number of fish characterized here and in the previous studies by others precludes a precise evaluation of the frequencies of the transposition events in these different transposon systems, we think that the Tol2 system is potentially useful as we could find fish with multiple insertions, which had not been reported in the studies using either the Tc3 or mariner element.
This study has provided the basis for developing novel transgenesis and insertional mutagenesis methods using the Tol2 transposon system in zebrafish. The next important steps toward the establishment of transgenesis and insertional mutagenesis methods using the Tol2 element are (i) to determine the frequency of transposition in the germ line more precisely and develop a protocol to increase the frequency, (ii) to determine whether integration of Tol2 can cause mutations efficiently, and (iii) to develop transposon vectors and put useful reporter genes into the vector. Efforts are in progress along these lines.
Acknowledgments
This work was supported by grants from the Japan Society for the Promotion of Science (to A.S.), Yamada Science Foundation, Ito Fish Science Foundation, and Inoue Foundation for Science, and by Grants-in-Aid from Japan Society for the Promotion of Science and the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviation
- kb
kilobase(s)
Footnotes
References
- 1.McClintock B. Proc Natl Acad Sci USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClintock B. Cold Spring Harbor Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Fedoroff N, Wessler S, Shure M. Cell. 1983;35:235–242. doi: 10.1016/0092-8674(83)90226-x. [DOI] [PubMed] [Google Scholar]
- 4.Pohlman R F, Fedoroff N V, Messing J. Cell. 1984;37:635–643. doi: 10.1016/0092-8674(84)90395-7. [DOI] [PubMed] [Google Scholar]
- 5.Müller-Neumann M, Yoder J I, Starlinger P. Mol Gen Genet. 1984;198:19–24. [Google Scholar]
- 6.Sutton W D, Gerlach W L, Schwartz D, Peacock W J. Science. 1984;223:1265–1268. doi: 10.1126/science.223.4642.1265. [DOI] [PubMed] [Google Scholar]
- 7.Döring H P, Tillmann E, Starlinger P. Nature (London) 1984;307:127–130. doi: 10.1038/307127a0. [DOI] [PubMed] [Google Scholar]
- 8.Heierhorst J, Lederis K, Richter D. Proc Natl Acad Sci USA. 1992;89:6798–6802. doi: 10.1073/pnas.89.15.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S. New Biol. 1992;4:382–388. [PubMed] [Google Scholar]
- 10.Radice A D, Bugaj B, Fitch D H A, Emmons S W. Mol Gen Genet. 1994;244:606–612. doi: 10.1007/BF00282750. [DOI] [PubMed] [Google Scholar]
- 11.Oosumi T, Belknap W R, Garlick B. Nature (London) 1995;378:672. doi: 10.1038/378672a0. [DOI] [PubMed] [Google Scholar]
- 12.Ivics Z, Izsvák Z, Minter A, Hackett P B. Proc Natl Acad Sci USA. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivics Z, Hackett P B, Plasterk R H, Izsvák Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 14.Tomita H. In: Medaka (Killifish), Biology and Strains. Yamamoto T, editor. Tokyo: Yugakusha; 1975. pp. 251–272. [Google Scholar]
- 15.Koga A, Inagaki H, Bessho Y, Hori H. Mol Gen Genet. 1995;249:400–405. doi: 10.1007/BF00287101. [DOI] [PubMed] [Google Scholar]
- 16.McGinnis W, Shermoen A W, Beckendorf S K. Cell. 1983;34:75–84. doi: 10.1016/0092-8674(83)90137-x. [DOI] [PubMed] [Google Scholar]
- 17.Streck R D, MacGaffey J E, Beckendorf S K. EMBO J. 1986;5:3615–3623. doi: 10.1002/j.1460-2075.1986.tb04690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sommer H, Carpenter R, Harrison B J, Saedler H. Mol Gen Genet. 1985;199:225–231. [Google Scholar]
- 19.Hehl R, Nacken W K F, Krause A, Saedler H, Sommer H. Plant Mol Biol. 1991;16:369–371. doi: 10.1007/BF00020572. [DOI] [PubMed] [Google Scholar]
- 20.Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H. Nature (London) 1996;383:30. doi: 10.1038/383030a0. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami K, Koga A, Hori H, Shima A. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- 22.Kawakami K, Shima A. Gene. 1999;240:239–244. doi: 10.1016/s0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- 23.Lin S, Gaiano N, Culp P, Burns J C, Friedmann T, Yee J-K, Hopkins N. Science. 1994;265:666–669. doi: 10.1126/science.8036514. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami K, Hopkins N. Trends Genet. 1996;12:9–10. doi: 10.1016/s0168-9525(96)90083-9. [DOI] [PubMed] [Google Scholar]
- 25.Stuart G W, McMurray J V, Westerfield M. Development (Cambridge, UK) 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- 26.Cantatore P, Roberti M, Pesole G, Ludovico A, Milella F, Gadaleta M N, Saccone C. J Mol Evol. 1994;39:589–597. doi: 10.1007/BF00160404. [DOI] [PubMed] [Google Scholar]
- 27.Koga A, Hori H. Genet Res. 1999;73:7–14. doi: 10.1017/s0016672398003620. [DOI] [PubMed] [Google Scholar]
- 28.Koga A, Suzuki M, Maruyama Y, Tsutsumi M, Hori H. FEBS lett. 1999;461:295–298. doi: 10.1016/s0014-5793(99)01479-9. [DOI] [PubMed] [Google Scholar]
- 29.Raz E, van Luenen H G A M, Schaerringer B, Plasterk R H A, Driever W. Curr Biol. 1997;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- 30.Fadool J M, Hartl D L, Dowling J. Proc Natl Acad Sci USA. 1998;95:5182–5186. doi: 10.1073/pnas.95.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long Q, Meng A, Wang H, Jessen J R, Farrell M J, Lin S. Development (Cambridge, UK) 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 32.Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 33.Gaiano N, Allende M, Amsterdam A, Kawakami K, Hopkins N. Proc Natl Acad Sci USA. 1996;93:7777–7782. doi: 10.1073/pnas.93.15.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Nature (London) 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 35.Allende M L, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- 36.Becker T S, Burgess S M, Amsterdam A H, Allende M L, Hopkins N. Development (Cambridge, UK) 1998;125:4369–4378. doi: 10.1242/dev.125.22.4369. [DOI] [PubMed] [Google Scholar]
- 37.Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawakami K, Amsterdam A, Shimoda N, Becker T, Mugg J, Shima A, Hopkins N. Curr Biol. 2000;10:463–466. doi: 10.1016/s0960-9822(00)00444-9. [DOI] [PubMed] [Google Scholar]