Abstract
We show that Drosophila expresses four genes encoding proteins with significant similarities with the thiolester-containing proteins of the complement C3/α2-macroglobulin superfamily. The genes are transcribed at a low level during all stages of development, and their expression is markedly up-regulated after an immune challenge. For one of these genes, which is predominantly expressed in the larval fat body, we observe a constitutive expression in gain-of-function mutants of the Janus kinase (JAK) hop and a reduced inducibility in loss-of-function hop mutants. We also observe a constitutive expression in gain-of-function Toll mutants. We discuss the possible roles of these novel complement-like proteins in the Drosophila host defense.
The Drosophila host defense has attracted considerable interest over the past years. Drosophila, like other insects, is able to mount a rapid and efficient response when challenged with various microorganisms. This response is prototypical for innate immune defenses and, because insects lack an adaptive response, it represents a valuable model for the study of ante-antibody immunity. The interest of the model is further underlined by the exceptional experimental possibilities offered by Drosophila molecular genetics.
Our current view of the Drosophila host defense is that septic injury activates proteolytic cascades, which lead to localized blood coagulation and melanization. Within 3 h, this activation is followed by the synthesis in the fat body, a functional equivalent of the mammalian liver, of several potent antimicrobial peptides, which are secreted into the hemolymph to oppose invading microorganisms. The induction of the genes encoding these peptides relies on intracellular signaling cascades that exhibit significant similarities with the activation of NF-κB in mammalian immune responses. Finally, circulating blood cells, the plasmatocytes, function as macrophages to engulf bacteria or fungal spores, and large-sized microorganisms are encapsulated by another blood cell-type, the lamellocyte (for recent reviews, see refs. 1 and 2).
Our information on the effector molecules of the Drosophila immune defense and the control of their expression has progressed significantly in the last decade. However, we are still largely ignorant as to the recognition of infectious non-self by the host. It has been proposed that germ-line-encoded pattern recognition receptors bind microbial cell wall determinants (such as lipopolysaccharides, mannans, and peptidoglycans) and initiate an immune response, either by activating associated proteases in circulation or by directly triggering intracellular signaling pathways in immune responsive cells (reviewed in refs. 1, 3, and 4). To date, no pattern recognition receptor has been firmly identified in Drosophila and shown to activate an immune response. Here, we have hypothesized that a primitive complement-like system, evocative of the alternative or the lectin pathways of complement, could be involved in the activation of some of the Drosophila host defense mechanisms. This hypothesis was made attractive by the recent reports that invertebrates such as sea urchins and tunicates have a complement-like system, and produce proteins with structural similarities to vertebrate complement C3 proteins, containing an intrachain thiolester bond (5, 6). Similar proteins have also been described in the horseshoe crab, a member of the class of arthropods to which also belongs Drosophila (7).
We now report the identification in the Drosophila genome of several genes coding for proteins that have the hallmark of the members of the superfamily of complement C3/α2-macroglobulins. We show that these genes are expressed in larvae and adults of Drosophila at a low level but, significantly, that their expression is up-regulated after an immune challenge. In the case of one of these genes, this up-regulation is clearly dependent on the Janus kinase hopscotch gene, which has already been variously implicated in immune responses in Drosophila and mammals (8, 9). We also observe a constitutive expression of this complement C3/α2-macroglobulin-like gene in a Toll gain-of-function mutant background (10, 11). Our data set the stage for a functional and genetic analysis of the involvement of complement-like proteins in the Drosophila innate immune response.
Materials and Methods
Computer Search and Sequence Analysis.
The Berkeley Drosophila Genome Project database (BDGP; http://www.fruitfly.org/) (12–14) was searched by using the α-chain of human complement C3 with the tblastn program. Subsequent DNA sequence analysis and comparison were performed by using Lasergen dnastar; protein analyses were carried out through different web servers such as the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nib.gov/) for general protein comparison; ExPASy (http://www.expasy.ch/tools/) and the Center for Biological Sequence analysis (CBS; http://www.cbs.dtu.dk/) for prediction of signal peptide cleavage sites and prosite scan; Baylor College of Medicine search launcher for general protein sequence/pattern searches (http://dot.imgen.bcm.tmc.edu:9331/).
Five sequences from three genomic P1 clones (13) were found to show similarities with the human complement C3 α-chain: DS00365 (hereafter referred to as thiolester-containing protein 1, Tep1); DS02501 (Tep2 and partial sequence for Tep3), and DS08491 (Tep4 and 5). Recently, the celera sequences CSC:AC020004 provided additional information on the genomic sequences of Tep3 (14). For Tep2-D, Tep2-E, and Tep3, partial sequences were available from the Berkeley Drosophila Genome Project (BDGP) expressed sequence tag (EST) database (BDGP/Howard Hughes Medical Institute (HHMI) EST Project, unpublished data). The following ESTs were purchased from Research Genetics (Huntsville, AL) and fully sequenced: GH08432 (Tep2-D), GH01829 (Tep2-E), and GH01146 (Tep3).
PCR and Sequencing Analysis.
Single-stranded cDNA (sscDNA) synthesis was performed on polyA-RNA extracted from embryos or immune-challenged larvae, by using an oligoT15 primer and SuperscriptII reverse transcriptase (GIBCO/BRL); the sscDNA was purified on a QIAquick spin PCR purification column (Qiagen, Chatsworth, CA). Putative intron–exon boundaries for the Tep1 and -4 genes were determined by comparing the overall deduced protein sequences with those of vertebrate complement C3 and α2-macroglobulin (blastx). Primers were designed from these predictions to produce double-stranded cDNA by PCR, by using sscDNA as a template, with the following protocol: 1 cycle at 95°C for 5 min; 30 cycles at 95°C for 0.5 min, 60°C for 0.5 min, and 72°C for 2 min; and 1 cycle at 72°C for 10 min. The following five sets of primers (forward and reverse) were used to cover the complete sequence of Tep1: gcaaaaaatattgcattatgctgtggttaa, tggaactcgatgcctccgctacc; gctaaaagttatgggtagcggag, tcgtagcttgaggcaccagtgca; tgcactggtgcctcaagctacga, ggccggattgagggctaagggaa; ttcccttagccctcaatccggcc, aatcgacaatcggtttgggttca; and cgaagtagagaccacctcttatg, tccgcactcgttgccgtggcaaat. For Tep4, the following sets were used: atccaatcaaaatgcgtcgcgca, acgactggagttgaggattactt; aagtaatcctcaactccagtcgt, cgcaacaggttctcaaggtt; and aaccttgagaacctgttgcg, ctctagcacttgctcttgcagtc. All cDNA fragments obtained were subcloned into pGEM-T easy vector (Promega) after purification on a QIAquick spin PCR purification column (Qiagen). Sequencing was performed by using the CEQ 2000 Dye Terminator Cycle Sequencing Kit (P/N608000) and a CEQ 2000 apparatus for analysis of DNA fragments by using capillary electrophoresis (Beckman Coulter).
For analysis of transcription, sscDNA templates were prepared from normal and immune-challenged wandering larvae or from various tissues of immune-challenged wandering larvae. The amount of sscDNA templates for each PCR was normalized with Rp49-specific primers (sense, atacaggcccaagatcgtga; antisense, acgttacagtgtattccgacc), after which 25 cycles of amplification were performed for Rp49 (nonsaturating conditions). For amplification of Tep1, 35 cycles were performed with Taq DNA polymerase (GIBCO/BRL) (1 cycle at 95°C for 5 min; 35 cycles at 95°C for 0.5 min, 60°C for 0.5 min, and 72°C for 2 min; and 1 cycle at 72°C for 10 min) with the following primers: sense, cgaagtagagaccacctcttatg; antisense, tccgcactcgttgccgtggcaaat.
Infection Procedure.
Bacterial challenge was performed by pricking third instar wandering larvae or 6-day-old adults with a fine needle dipped into a concentrated culture of Escherichia coli and Micrococus luteus.
RNA Extraction and Northern Blot Analysis.
Total RNA were extracted with Trizol reagent (GIBCO/BRL), and polyA-RNA was purified by oligotex beads (Qiagen) according to the supplier's protocols. For Northern blot analysis, polyA-RNA (5 μg per sample) was fractionated by denaturating gel electrophoresis in 0.7% agarose/formaldehyde gels with Mops buffer (25 mM Mops/5 mM sodium acetate/2 mM EDTA). After transfer to a nylon membrane, the RNAs were hybridized with random-primed cDNA probes (Rediprime II, Amersham; [32P]dCTP, 3000 Ci/mmol) overnight at 42°C by using classical Denhardt's hybridization buffer containing 50% formamide (15). Rp49 cDNA served as internal control (16).
Drosophila Stocks and Genetic Crosses.
Flies were maintained at 20°C-22°C on a standard cornmeal medium. The mutant strains were as described earlier: imd (17), Toll10B (18), Toll9QRE (19), hoptum-l (20, 21), hopM38 (22), and spzrm7 (18). Appropriate balancers marked by actGFP (23) or UbiGFP (C. Thummel, unpublished data) were used to identify homozygous mutant larvae. To generate hopM38;Toll10B double mutants, hopM38/FM7actGFP females were crossed to Toll10B/TM6UbiGFP males, and male larvae not carrying the green fluorescent protein (GFP) balancers were selected from the progeny.
Results
Drosophila Possesses Five Distinct Genes Encoding Complement C3/α2-Macroglobulin-Like Proteins.
We first asked whether the Drosophila genome encoded sequences corresponding to complement-like proteins. For this, we conducted a blast search of the Berkeley Drosophila Genome Sequencing Project Database with the amino acid sequence of the α-chain of human complement C3. This search produced several hits. Relevant sequences were present on three P1 clones (13) and correspond to five distinct genes. Four of these putative genes are transcribed, whereas we were not able to amplify any cDNA corresponding to the fifth putative gene, which possibly corresponds to a pseudogene. We complemented the information on the intron–exon boundaries of the four interesting genes by sequencing full-length cDNAs found in EST libraries or by amplifying cDNA fragments from immune-challenged larvae and adults (Fig. 1, and Material and Methods). We next analyzed by the clustal-pam method the protein sequences deduced from the cDNAs, which confirmed that these molecules exhibit significant similarities with the thiolester-containing proteins of the complement C3/α2-macroglobulin superfamily. Pending a detailed functional analysis, we will refer to these proteins as thiolester-containing proteins (TEPs), and number them from 1 to 4 for the expressed genes (see accession numbers in footnote). We will reserve the number 5 to the fifth DNA sequence. Note that an additional similar cDNA sequence had been previously described by T. Crowley et al. (unpublished results; sequence NCBI accession number: Y11116). This complement-like protein lacks the thiolester motif and will not be considered hereafter.
Figure 1.
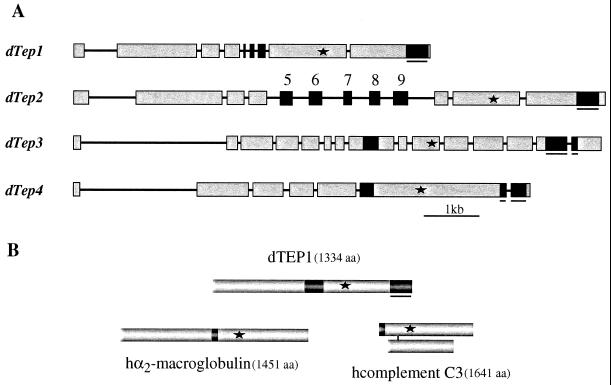
(A) Gene organization of Drosophila Tep1, -2, -3, and -4. Exons are represented by boxes and introns by lines. Asterisks indicate the positions of the thiolester motifs. Black nonunderlined boxes correspond to the variable region of TEPs. Five isoforms of Tep2 transcripts have been isolated; each transcript differs only by a single exon (exons 5 to 9) (E.P. and M.L., unpublished data). Black underlined boxes correspond to the highly conserved C-terminal region containing six cysteines. Chromosome localizations are as follows: Tep1, 35F1-F4; Tep2 and 3, 28B1-B4 (these genes are positioned at 2 kb from one another in reverse orientation); Tep4, 37F1-F2. (B) Protein structures of the three subfamilies of thiolester-containing proteins represented by Drosophila TEP1, human α2-macroglobulin, and human complement C3. The asterisks indicate the positions of the thiolester motifs, the black boxes show the respective positions of the variable domain in TEP, the bait domain of α2-macroglobulin, and the anaphylatoxin of complement C3. The underlined black box corresponds to the particularly well conserved region in TEPs (see Fig. 2).
The deduced protein sequences indicate that TEPs 1 to 4 have a putative signal peptide and hence are likely to correspond to secreted proteins. TEPs 1 to 4 show significant similarities (e.g., 49% sequence identities between TEP1 and TEP2). The four sequences share three regions that are noteworthy: (i) all TEPs contain a highly conserved region of 30-aa residues harboring a canonical thiolester motif GCGEQ (see asterisk in Fig. 1, and supplementary Fig. 6, File 1, block G, which is published as supplemental data on the PNAS web site, www.pnas.org); (ii) the C-terminal parts of the four deduced proteins are highly conserved over a 126-aa stretch and contain six cysteines in conserved positions (Fig. 2); and (iii) all TEPs show a highly variable central region (e.g., between residues 581 and 642 in TEP1). Interestingly, this highly variable region overlaps in TEP2 with the position of exons 5 to 9, which undergo alternative splicing as evidenced by cDNA cloning studies (see Fig. 1).
Figure 2.
Alignment by the clustal method of the highly conserved C-terminal region of Drosophila TEP1 (amino acid residues 1230 to 1354), TEP2 (1280 to 1404), TEP3 (1299 to 1422), and TEP4 (1375 to 1497). Identical residues are in bold. Cysteines are underlined. This region confers the specific signature of TEPs within the complement C3/α2-macroglobulin superfamily.
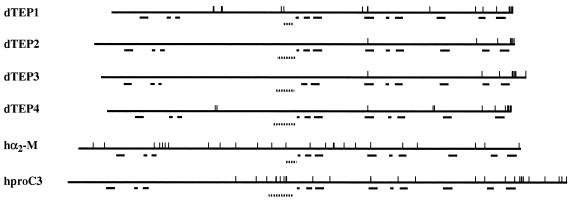
The sequence similarities between TEPs and the proteins of the complement C3/α2-macroglobulin superfamily are distributed all over the proteins. In vertebrates, all members of this superfamily are characterized by the presence of 12 blocks of moderately conserved residues in similar relative positions (block search; ref. 24) and are referred to as blocks A to L. Remarkably, with minor variations, the 12 blocks defining the signature of this superfamily are also present in the Drosophila TEPs (Fig. 3; see also File 1, supplementary Fig. 6). In vertebrates, block D is located C-terminally to the less conserved region of the superfamily, corresponding to the bait domain in α2-macroglobulin and to the anaphylatoxin domain in complement C3. Interestingly, in TEPs, the equivalent of block D is located C-terminally to the highly variable region, which we have described above. This block contains N-terminally a putative proteolytic cleavage site (RK) present in all TEPs (see File 1, supplementary Fig. 6). A characteristic feature of vertebrate thiolester proteins is a relatively high number of cysteines whose pattern is different in α2-macroglobulins and complement C3 (25). The numbers of cysteines is markedly lower in TEPs, and no clear-cut alignment is apparent with the patterns described for α2-macroglobulins or for complement C3. Conversely, the characteristic cysteine signature of the C-terminal region common to all TEPs (Fig. 2) is absent from the vertebrate proteins (see Figs. 1 and 3).
Figure 3.
Schematic representation of the protein structure of Drosophila TEP1 to -4, human α2-macroglobulin, and the proprotein for human complement C3; vertical bars show the positions of cysteine residues; horizontal bars indicate the positions of the various domain-signatures defined for the superfamily of thiolester-containing proteins. From left to right, blocks A to L (see also file 1, supplementary Fig. 6). Dotted lines show the positions of hypervariable regions in TEPs, the position of the bait domain of α2-macroglobulin, and anaphylatoxin in complement C3.
Transcriptional Profiles of Tep Genes.
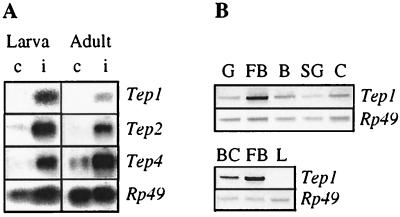
To investigate the expression patterns of the Tep genes, we probed Northern blots with the appropriate cDNAs. Tep1, -2, -3, and -4 showed a low basal level of expression during larval, pupal, and adult stages (data not shown). Interestingly, the expression of the Tep1, -2, and -4 genes was strongly up-regulated after bacterial challenge in larvae (Fig. 4A), which was particularly evident 6 h after infection. Expression of Tep2 and Tep4 was also markedly induced by challenge in adults, whereas that of Tep1 was more discrete at this stage (Fig. 4A). Finally, the Tep3 gene expression was not noticeably modified by immune challenge, either in larvae or in adults.
Figure 4.
(A) Transcription levels of Drosophila Tep1, -2, and -4 before (c) and after (i) an immune challenge (6 h) in wandering larvae, and in 6-day-old adult wild-type Drosophila. Five micrograms of polyA-RNA were fractionated by electrophoresis in denaturating agarose-formaldehyde gels. After transfer to a nylon membrane, the RNAs were hybridized with random-primed [32P]cDNA probes corresponding to Tep1, -2, and -4; and Rp49 for the loading control. (B) Transcription level of Tep1 in different tissue extracts of immune-challenged larvae: G, gut; F, fat body; B, brain; SG, salivary glands; C, carcass; BC, blood cells; and L, l(2)mbn cells challenged with bacteria. DNA fragments were amplified by PCR with specific primers of Tep1 and separated on agarose gel. The Rp49 amplification product served as an internal control.
In the following, we have focused our attention on the control of transcription of the Tep1 gene. This gene yields a single transcript that is strongly up-regulated, whereas Tep2 yields at least five splice isoforms. As will become apparent below, Tep1 is also the strongest reactant in hop mutants and provided the most convenient tool in this context.
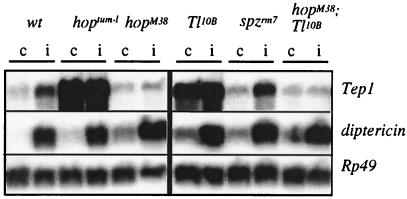
Tep1 is mainly transcribed in the fat body, as illustrated by the comparison of intensities of PCR amplification products from RNA extracts of gut, fat body, brain, salivary gland, and body wall (Fig. 4B). We also observed a marked expression in blood cells collected from immune-challenged larvae, but not in challenged (incubation with lipopolysaccharide, bacteria, or fungal spores) l(2)mbn or S2 Schneider cell lines (Fig. 4B). This latter result was somehow surprising because these cells are generally considered as hemocyte-derived (26). As stated in the introduction, the fat body of Drosophila is a major immune-responsive tissue and strongly expresses antimicrobial peptide genes on immune challenge. Two signaling pathways regulate this induction: the Toll pathway is primarily responsible for expression of the antifungal peptide gene drosomycin, whereas the imd pathway controls expression of most antibacterial peptide genes (for reviews, see refs. 1, 10, and 11). Because Tep1 is predominantly expressed in the fat body (Fig. 4B), we have first analyzed whether one or both of these pathways also controlled Tep1 induction after immune challenge. We have first addressed imd mutants and have not observed any difference in induction of Tep1 in this background, as compared with wild-type larvae (data not shown). In Toll gain-of-function mutants, we observed a strong constitutive expression of Tep1 (Fig. 5). However, in loss-of-function mutants, for the gene encoding the Toll-ligand Spaetzle, the level of Tep1 induction by immune challenge was normal. We also addressed Toll loss-of-function mutant larvae. Only a small number of these fragile larvae could be submitted to immune challenge, and Tep1 was up-regulated as in wild-type (data not shown). These observations suggest that the constitutive expression of Tep1 observed in Toll gain-of-function mutants is an indirect effect.
Figure 5.
Transcription levels of Drosophila Tep1 before (c) and after (i) immune challenge (6 h) in wandering larvae in different genetic backgrounds: wt (wild-type), hoptum-l, hopM38, Toll10B, spzrm7, and hopM38;Toll10B double mutants. Five micrograms of polyA-RNA were fractionated by electrophoresis in denaturating agarose-formaldehyde gels. After transfer to a nylon membrane, the RNAs were hybridized with random-primed cDNA probes: Tep1, Rp49, and diptericin. The Rp49 cDNA probe served as an internal control for quantification of RNA, whereas the diptericin probe served as a control for an efficient immune challenge of insects.
Toll overexpression leads to overproliferation and abnormal differentiation of the blood cells in Drosophila (27). It has been suggested that this effect is mediated by the JAK kinase hopscotch (hop) (8, 27). We examined expression of Tep1 in a gain-of-function mutant of hop (hoptum-l) and, remarkably, observed a strong constitutive expression of the Tep1 gene. The role of hop in the expression of Tep1 was corroborated by the observation that, in loss-of-function hopM38 mutants, Tep1 induction by septic injury was dramatically reduced as compared with wild-type (Fig. 5). We have further generated hopscotch loss-of-function;Toll gain-of-function, double mutants. As illustrated in Fig. 5, the Toll-driven expression of Tep1 was abolished in a hop loss-of-function background.
Discussion
The data presented above show that Drosophila expresses at least four distinct proteins that contain a canonical thiolester motif and exhibit overall stringent similarities with the thiolester-containing proteins of the complement C3/α2-macroglobulin superfamily. Importantly in the context of the present study, the genes encoding these proteins are up-regulated by immune challenge and this process is mediated, at least for one of the proteins, TEP1, by the hop gene already known to be involved in the regulation of immune responses in Drosophila. Also relevant is the observation that the protein sequences deduced from the Tep genes contain putative signal peptides, indicating that they correspond to secreted proteins. Although the Drosophila thiolester-containing proteins are clearly related to the superfamily of complement C3/α2-macroglobulin, a certain number of structural characteristics, and namely the very distinct numbers and positions of cysteines, support the view that TEPs form a separate group of thiolester proteins that either have evolved independently, or have separated very early from the hypothetical common ancestor molecule of complement C3/α2-macroglobulins.
In higher vertebrates, the complement system consists of about 30 serum and cell surface proteins and mediates inflammatory reactions, opsonization of microorganisms for phagocytosis, and direct killing of some pathogens. Activation can occur via the classical antibody-dependent pathway, the alternative pathway, and the lectin pathway, which all converge on the central complement C3 protein (for recent reviews, see refs. 28 and 29). The presence in Drosophila of several proteins with basic structural characteristics similar to complement C3 makes the working hypothesis attractive that an ancient equivalent of the alternative pathway and/or the lectin pathway exists in this species. In vertebrates, the activation of the alternative pathway is initiated by spontaneous hydrolysis of the thiolester bond of complement C3, resulting, through association with other proteins of the complement system, in an active C3 convertase that is normally inactivated by regulatory proteins present on self tissue, but absent from non-self, providing for a relative primitive mode of discriminating self from non-self. Active C3 convertase in turn activates complement C3 and, through an amplification loop, triggers the conventional effector mechanisms of complement. Activation of the lectin pathway is initiated when various sugars present on the surface of microorganisms bind to a collectin, the mannan-binding lectin (MBL), thereby inducing proteolytic cascades that activate complement C3 and the downstream events common to all three activation pathways.
In the mid-nineties, it became apparent that the complement system is not a unique property of the host defense armatarium of vertebrates. ESTs from cDNA libraries of sea urchin coelomocytes were found to encode a protein with structural similarities to vertebrate complement C3, including an intrachain thiolester motif, plus a homologue of vertebrate factor B, which participates in the activation of complement C3 through the alternative pathway (28). More recently, an ascidian species was reported to possess homologues of complement C3 and of two mannose-binding lectin-associated proteases (MASPs), plus a homologue of factor B, raising the possibility that equivalents of both the lectin and the alternative activation pathways are present in these deuterostome invertebrates (28). Experiments with ascidian coelomocytes further indicate that the complement C3-like molecules act as opsonic factors and are activated through a complement-like cascade (28).
TEPs are also structurally close to α2-macroglobulins, which are evolutionary ancient protease inhibitors, from which complement C3 has been proposed to have arisen by gene duplication (reviewed in ref. 30). Protease inhibitors related to α2-macroglobulin have been described in several invertebrates and were particularly well studied in the horseshoe crab Limulus (7, 31). Indeed, Limulus α2-macroglobulin has been proposed to function as a protease inhibitor, particularly of proteases released by tissue damage caused by injury or pathogens and of soluble or surface bound proteases produced by invading microorganisms (32). Dodds and Law (30) suggest that the first opsonic system could have required no specific recognition or activation mechanism other than the presence of exogenous proteases causing α2-macroglobulin to bind directly to the protease-producing organism.
Because Drosophila has four expressed genes encoding proteins with structural similarities to the superfamily of complement C3/α2-macroglobulin, we may expect significant functional versatilities, all the more so, because one of the Tep genes, Tep2, gives rise to five different transcripts. Interestingly, the Tep2 transcripts are identical except for a short region of 30 aa. This region is encoded by alternatively spliced exons corresponding to the hypervariable region of the TEPs; it is located in a relative position similar to the bait domain in α2-macroglobulins or the anaphylatoxins in complement C3. Alternative splicing has not been reported in vertebrates for members of the complement C3/α2-macroglobulin superfamily. By increasing the number of putative recognition motifs for microorganisms or proteases, it may contribute to the fine-tuning of recognition of noxious structural patterns in the absence of the large repertoire of receptors of the adaptive immune response in vertebrates.
The Drosophila plasmatocytes are macrophage-like blood cells that readily engulf bacteria or fungal spores, as well as various cellular debris resulting from injury or apoptosis. Nothing is known about possible opsonization in this model, and a tempting working hypothesis is that the complement-like proteins that we describe here precisely fulfill such a role in the host defense. Our future efforts will be directed toward experimentally testing this hypothesis, and we anticipate that the generation of mutants of the various Tep genes, which all map to the left arm of the second chromosome, will be invaluable in this endeavor.
TEP1 is produced mainly in the fat body, and its expression is up-regulated by immune challenge. We hypothesized that, as is the case for immune-induction of antimicrobial peptides in this tissue, the up-regulation would be dependent on either the Toll or the imd pathway. We were highly interested to note that this control is strongly dependent on the JAK hopscotch. Hopscotch is the only JAK identified in Drosophila, and in the gain-of-function mutant hopTum-l, Tep1 is constitutively expressed, whereas its immune-inducibility is dramatically reduced in the loss-of-function mutant hopM38. Gain-of-function mutations of hop have remarkable effects on hemopoiesis in Drosophila and result in overproliferation of blood cells, increased differentiation of lamellocytes, and aggregation of blood cells into masses, which tend to become melanized, a process referred to as melanotic tumor formation. Our data thus show that these events are concomitant with an increased transcription of the Tep1 gene. Whether they are causally related remains an open question, but it will be worthwhile investigating whether the TEP1 protein can affect the aggregation of blood cells and the localized induction of melanization.
In addition to its established role in the control of synthesis of the antifungal peptide drosomycin, the Toll pathway has been proposed by Govind and associates (27) to be implicated in the control of hemocyte density. Hemocyte numbers are increased in Toll gain-of-function mutants. Of potential interest in the present context is the observation that, in these mutants, melanotic tumors develop that are similar to those seen in JAK gain-of-function mutants. We have also observed that, in Toll gain-of-function mutants, Tep1 is strongly expressed in the absence of immune challenge. However, we believe that this effect must be indirect, because immune challenge can up-regulate Tep1 expression in spaetzle and Toll loss-of-function mutants. Toll gain-of-function mutants are known to produce a large number of peptides or polypeptides, referred to as Drosophila immune-induced molecules (DIMs) (ref. 33; P. Bulet and L. Sabatier, personal communication), which are absent from wild-type nonchallenged insects. These and/or other modifications in the hemolymph induced by melanotic tumors might account for the constitutive expression of Tep1 in Toll gain-of-function mutants.
Supplementary Material
Acknowledgments
We are indebted to Dr. Charles Dearolf (Massachusetts General Hospital, Boston) and the Bloomington Stock center for sending fly stocks. The technical assistance of Marie-Eve Moritz and Claude Chevalier is gratefully acknowledged. This study was supported by Institutional funds from Centre National de la Recherche Scientifique, the University Louis Pasteur of Strasbourg, and National Institutes of Health Grant 5P01AI44220-02.
Abbreviations
- TEP
thiolester-containing protein
- JAK
Janus kinase
- EST
expressed sequence tag
- sscDNA
single-stranded cDNA
- hop
hopscotch
Footnotes
References
- 1.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 2.Meister M, Hetru C, Hoffmann J A. Curr Top Microbiol Immunol. 1999;248:17–36. doi: 10.1007/978-3-642-59674-2_2. [DOI] [PubMed] [Google Scholar]
- 3.Janeway C. Cold Spring Harbor Symp Quant Biol. 1989;54:1–13. [PubMed] [Google Scholar]
- 4.Medzhitov R, Janeway C A., Jr Semin Immunol. 1998;10:351–353. doi: 10.1006/smim.1998.0136. [DOI] [PubMed] [Google Scholar]
- 5.Al-Sharif W Z, Sunyer J O, Lambris J D, Smith L C. J Immunol. 1998;160:2983–2997. [PubMed] [Google Scholar]
- 6.Courtney Smith L, Azumi K, Nonaka M. Immunopharmacology. 1999;42:107–120. doi: 10.1016/s0162-3109(99)00009-0. [DOI] [PubMed] [Google Scholar]
- 7.Iwaki D, Kawabata S, Miura Y, Kato A, Armstrong P B, Quigley J P, Nielsen K L, Dolmer K, Sottrup-Jensen L, Iwanaga S. Eur J Biochem. 1996;242:822–831. doi: 10.1111/j.1432-1033.1996.0822r.x. [DOI] [PubMed] [Google Scholar]
- 8.Mathey-Prevot B, Perrimon N. Cell. 1998;92:697–700. doi: 10.1016/s0092-8674(00)81396-3. [DOI] [PubMed] [Google Scholar]
- 9.Dearolf C R. Cell Mol Life Sci. 1999;55:1578–1584. doi: 10.1007/s000180050397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson K V. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 11.Imler J-L, Hoffmann J A. Curr Opin Microbiol. 2000;3:16–22. doi: 10.1016/s1369-5274(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 12.Rubin G M. Genome Res. 1996;6:71–79. doi: 10.1101/gr.6.2.71. [DOI] [PubMed] [Google Scholar]
- 13.Kimmerly W, Stultz K, Lewis S, Lewis K, Lustre V, Romero R, Benke J, Sun D, Shirley G, Martin C, et al. Genome Res. 1996;6:414–430. doi: 10.1101/gr.6.5.414. [DOI] [PubMed] [Google Scholar]
- 14.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.O'Connell P, Rosbach M. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J, Hoffmann J. Proc Natl Acad Sci USA. 1995;92:9365–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaitre B, Nicolas E, Michaut L, Reichhart J, Hoffmann J. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 19.Anderson K V, Jürgens G, Nüsslein-Volhard C. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- 20.Hanratty W P, Ryerse J S. Dev Biol. 1981;83:238–249. doi: 10.1016/0012-1606(81)90470-x. [DOI] [PubMed] [Google Scholar]
- 21.Luo H, Hanratty W, Dearolf C. EMBO J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrimon N, Mahowald A P. Dev Biol. 1986;118:28–41. doi: 10.1016/0012-1606(86)90070-9. [DOI] [PubMed] [Google Scholar]
- 23.Reichhart J M, Ferrandon F. Drosophila Inf Serv. 1998;81:201–202. [Google Scholar]
- 24.Henikoff S, Henikoff J G. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 25.Sottrup-Jensen L, Stepanik T M, Kristensen T, Lonblad P B, Jones C M, Wierzbicki D M, Magnusson S, Domdey H, Wetsel R A, Lundwall A, et al. Proc Natl Acad Sci USA. 1985;82:9–13. doi: 10.1073/pnas.82.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gateff E. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- 27.Qiu P, Pan P C, Govind S. Development. 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- 28.Nonaka M, Azumi K, Ji X, Namikawa-Yamada C, Sasaki M, Saiga H, Dodds A W, Sekine H, Homma M K, Matsushita M, et al. J Immunol. 1999;162:387–391. [PubMed] [Google Scholar]
- 29.Prodinger W M, Würzner R, Erdei A, Dierich M P. In: Fundamental Immunology. Paul E B W E, editor. Philadelphia: Lippincott-Raven; 1999. pp. 967–995. [Google Scholar]
- 30.Dodds A W, Law S K. Immunol Rev. 1998;166:15–26. doi: 10.1111/j.1600-065x.1998.tb01249.x. [DOI] [PubMed] [Google Scholar]
- 31.Enghild J J, Thogersen I B, Salvesen G, Fey G H, Figler N L, Gonias S L, Pizzo S V. Biochemistry. 1990;29:10070–10080. doi: 10.1021/bi00495a009. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong P B, Melchior R, Swarnakar S, Quigley J P. Mol Immunol. 1998;35:47–53. doi: 10.1016/s0161-5890(98)00007-8. [DOI] [PubMed] [Google Scholar]
- 33.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann J A, Bulet P. Proc Natl Acad Sci USA. 1999;95:11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.