Abstract
We have identified a type I cytokine receptor, which we have termed novel interleukin receptor (NILR), that is most related to the IL-2 receptor β chain (IL-2Rβ) and physically adjacent to the IL-4 receptor α chain gene on chromosome 16. NILR mRNA is most highly expressed in thymus and spleen, and is induced by phytohemagglutinin in human peripheral blood mononuclear cells. NILR protein was detected on human T cell lymphotropic virus type I-transformed T cell lines, Raji B cells, and YT natural killer-like cells. Artificial homodimerization of the NILR cytoplasmic domain confers proliferation to Ba/F3 murine pro-B cells but not to 32D myeloid progenitor cells or CTLL-2 murine helper T cells. In these latter cells, heterodimerization of IL-2Rβ and the common cytokine receptor γ chain (γc) cytoplasmic domains allows potent proliferation, whereas such heterodimerization of NILR with γc does not. This finding suggests that NILR has signaling potential but that a full understanding of its signaling partner(s) is not yet clear. Like IL-2Rβ, NILR associates with Jak1 and mediates Stat5 activation.
Type I cytokine receptors represent a class of receptors, including those for many interleukins such as IL-2 to IL-7, IL-9, IL-11 to IL-13, and IL-15, as well as other cytokines, such as granulocyte–macrophage colony-stimulating factor, oncostatin M, ciliary neurotrophic factor, cardiotrophin-1, growth hormone, prolactin, erythropoietin, and thrombopoietin (1, 2). Many of these cytokines play important roles related to the development or function of lymphohematopoietic lineages and sometimes share common components. For example, IL-3, IL-5, and granulocyte–macrophage colony-stimulating factor exert actions on hematopoietic cells and share a common β chain (3), whereas IL-2, IL-4, IL-7, IL-9, and IL-15 act on lymphocytes and share the common cytokine receptor γ chain, γc (2, 4). Mutation of γc results in X-linked severe combined immunodeficiency in humans, a disease characterized by an absence of T cells and natural killer (NK) cells, and nonfunctional B cells (5, 6). Identification of additional type I cytokine receptors thus can elucidate biological systems and potentially advance our understanding of molecular mechanisms of human disease. We now report the identification of a type I cytokine receptor that is most related to the IL-2 receptor β chain (IL-2Rβ). The gene encoding this receptor is immediately adjacent to the gene encoding the IL-4 receptor α chain on chromosome 16p12. Expression is most abundant in lymphoid tissues and is induced by stimuli that act through the T cell antigen receptor, suggesting that this receptor mediates signals important for the immune system.
Materials and Methods
Gene Prediction and Analysis.
The human high-throughput genomic sequence was scanned for “virtual” ORFs by using the ab initio gene prediction program genscan (7). Comparison of predicted ORFs to known proteins was performed by using blast (8). The signal sequence was predicted by using the signalp algorithm (9). Transmembrane predictions were performed with tmpred (http://www.ch.embnet.org/software/TMPRED_form.html), an algorithm based on the statistical analysis of a transmembrane domain database, as well as based on an analysis of transmembrane domains (10). Amino acid alignment was performed by using clustal w (11).
Isolation of a Novel Interleukin Receptor (NILR).
To isolate the cDNA for NILR, nested PCR primers were designed around the putative start and stop codons, and PCR was performed on Marathon cDNA (CLONTECH) from bone marrow, fetal liver, leukocyte, placenta, spleen, and lung. The 5′ primers were 5′-CAAGTCTGCCTGGGAAGAGACAGGATG-3′ and nested 5′-GCCTGGCTCACCCTCCACTGTACGTCTCTT-3′. The 3′ primers were 5′-ACCAAAGGCTGCAGGTGTCTTCACATCACA-3′ and nested 5′-CACAGCCCAGGTGACCTTGTCTCTGGCTCA-3′. The PCR products were TA cloned (Invitrogen) and sequenced.
A cDNA library in λZAP-II prepared from human YT NK-like cells was screened by using the nested PCR product from bone marrow. From 5 × 105 phage clones, 15 clones were identified after tertiary screening and phagemids were excised by using an Excision kit (Stratagene). Two cDNAs contained the full-length NILR-coding region. Sequencing was done with an Applied Biosystems model 310 DNA sequencer.
Murine cDNA Isolation and Cloning.
We screened an adult murine thymus cDNA library (provided by Paul Love, National Institute of Child Health and Human Development) by using a human NILR probe. Of nine different human probes tested on mouse genomic DNA, the nucleotide 471–830 probe (numbering scheme of Fig. 1) gave the strongest cross-species hybridization and was used. Hybridization was done overnight with QuikHyb (Stratagene). Membranes were washed in 2× SSC/0.1% SDS at 45°C for 15 min twice, and then in 0.2× SSC/0.1% SDS at 60°C for 30 min.
Figure 1.
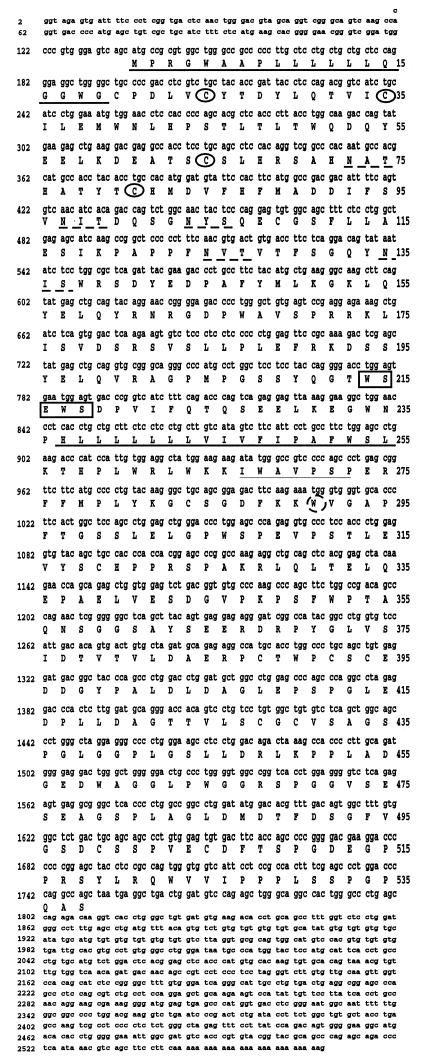
NILR encodes a type I cytokine receptor. DNA and predicted amino acid sequences of human NILR cDNA clone 17. Conserved Cys residues are circled. The predicted signal peptide and transmembrane domains are underlined. The boundaries of the transmembrane domain are as predicted by ref. 10; tmpred instead predicts that the domain begins at Leu-238 instead of at His-237. The potential N-linked glycosylation sites are underlined with broken lines. The WSXWS motif is boxed. Thin underline and broken circles denote the Box B1 region and conserved Trp, respectively.
After obtaining a partial murine NILR cDNA, we made a murine NILR probe corresponding to the fourth coding exon and used this to screen the same library as well as a cDNA library prepared from murine splenocytes stimulated with Con A. We identified a murine cDNA that lacked the 5′ end (first two coding exons). The 5′ end of the cDNA was isolated by using mouse spleen Marathon cDNA and 5′ rapid amplification of cDNA ends (RACE) according to the manufacturer's instructions (CLONTECH).
Cell Lines.
MT-2 and HUT-102B2 are human T lymphotrophic virus type I (HTLV-I)-transformed T cell lines; YT is a NK-like cell line. Molt-4, CEM, and Jurkat are T cell acute lymphocytic leukemia cell lines; Raji is an Epstein–Barr virus-transformed Burkitt lymphoma cell line. K562 is an erythroleukemia cell line. These cells were maintained in RPMI medium 1640 supplemented with 10% FBS/2 mM l-glutamine/penicillin/streptomycin (RPMI complete medium). 293T cells were maintained in DMEM containing 10% FBS/glutamine/penicillin/streptomycin. Ba/F3 is a murine IL-3-dependent pro-B cell line, CTLL-2 is a murine IL-2-dependent helper T cell line, and 32D is a murine IL-3-dependent myeloid cell line. Ba/F3 and 32D cells were grown in RPMI complete medium containing 5 × 10−5 M 2-mercaptoethanol and 5% WEHI-3B-conditioned medium as a source of IL-3. CTLL-2 cells were grown in the same medium except that WEHI-3B-conditioned medium was replaced by 50 units/ml IL-2.
Northern Blotting.
The mouse 12-tissue poly(A)+ RNA Northern blot membrane was from OriGene Technologies (Rockville, MD). The murine NILR probe was made by SmaI digestion of the pCI-murine NILR expression vector, and corresponded to coding region nucleotides 8–981 (numbered from ATG; nucleotides 1–3). Poly(A)+ RNA was prepared from human peripheral blood mononuclear cells (PBMC) stimulated with 2 μg/ml phytohemagglutinin-L (PHA) (Boehringer Mannheim); 0.8 μg of poly(A)+ RNA was used for each lane. Blots were hybridized with the same probe that was used for screening the human cDNA library or a control probe (0.3-kb pHe7 cDNA) (12). Hybridization was performed in 5× SSPE (1× SSPE is 0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA)/50% formamide/0.2% SDS/5× Denhardt's solution (1× Denhardt's solution is 0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA) at 42°C and washed in 2× SSPE/0.1% SDS for 15 min twice at 42oC, and then in 0.1× SSPE/0.1% SDS for 30 min at 55°C.
Production of Anti-NILR Antiserum.
To prepare antisera recognizing the cytoplasmic domain of NILR, we made a glutathione S-transferase (GST)-fusion protein corresponding to the cytoplasmic domain of NILR. Antisera were raised in goats by standard methods (Duncroft, Lovettsville, VA).
Western Blotting and Immunoprecipitation.
Anti-Jak1 polyclonal antiserum was used for immunoprecipitation (Santa Cruz Biotechnology), and a mAb was used for Western blotting (BD Transduction Laboratories, San Diego). Anti-IL-2Rβ and γc polyclonal antisera were from Santa Cruz Biotechnology. Anti-phosphotyrosine mAb 4G10 was purchased from Upstate Biotechnology (Lake Placid, NY). Anti-FLAG M2 mAb was from Sigma. A Stat5 polyclonal antiserum (13) was used for immunoprecipitation and a mAb (BD Transduction Laboratories) was used for Western blotting. Cell starvation was performed by washing cells and incubating them in RPMI complete medium 1640 without growth factors for 4 h. Cells were then stimulated for 10 min with 20 units/ml human recombinant erythropoietin (EPO; provided by Harvey Lodish, Whitehead Institute for Biomedical Research, Cambridge, MA). Cells were lysed with lysis buffer [50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.5% Nonidet P-40/1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF)/2 μg/ml leupeptin/2 μg/ml aprotinin/1 mM Na3VO4] on ice for 15 min, nuclei were centrifuged at 13,000 × g for 15 min, and lysates were immunoprecipitated with antibody and protein A-Sepharose (Amersham Pharmacia) for 1.5 h, washed, and subjected to SDS/PAGE. After transfer to Immobilon-P membranes (Millipore), immunoblotting was performed with enhanced chemiluminescence (Pierce).
Expression in 293T Cells.
Transfection of 293T cells was performed by the calcium phosphate method (5 Prime → 3 Prime). Subconfluent cells were diluted to 1:10–15 and transfected the next day with 5 μg of DNA.
Chimeric Receptor Proliferation Assay.
The retroviral vectors LXSN and LXSH (provided by Dustin Miller, Fred Hutchinson Cancer Research Center, Seattle) were used to transduce Ba/F3, 32D, and CTLL-2 cells with chimeric constructs containing the extracellular domain of the EPO receptor (EPOR) and the transmembrane and cytoplasmic domains of NILR, IL-2Rβ, or γc (EPOR/NILR, EPOR/IL-2Rβ, and EPOR/γc). The transmembrane and cytoplasmic domains of NILR, IL-2Rβ, and γc were generated by PCR and inserted after the NheI site near the 3′ end of the EPOR extracellular domain. After drug selection with G418 (Geneticin, Life Technologies, Gaithersburg, MD; 800 μg/ml for 7 days) and hygromycin (CLONTECH; 1,200 μg/ml for 7 days), the expression of chimeric receptors was confirmed by immunoprecipitation and Western blotting. Ba/F3 or 32D transfectants (2,000 cells) and CTLL-2 transfectants (4,000 cells) were plated in 96-well plates in medium alone, 100 units/ml EPO, 5% WEHI-3B-conditioned medium, or 100 units/ml IL-2. [3H]Thymidine was added 4 days later and incorporation was measured after 4 h. Experiments were performed in triplicate.
Electrophoretic Mobility-Shift Assays.
Cells were lysed in low salt buffer (10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EGTA/0.1 mM EDTA/1 mM DTT/1 mM AEBSF/2 μg/ml leupeptin/2 μg/ml aprotinin/1 mM Na3VO4) and centrifuged, and nuclear extracts were prepared with high salt buffer (20 mM Hepes, pH 7.9/0.4 M NaCl/1 mM EGTA/1 mM EDTA/1 mM DTT/1 mM AEBSF/2 μg/ml leupeptin/2 μg/ml aprotinin/1 mM Na3VO4) with agitation. Nuclear extracts (15–30 μg) were incubated with 20,000 cpm of a double-stranded β-casein promoter GAS (γ-interferon activated sequence) sequence (5′-AGATTTCTAGGAATTC-3′) probe for 30 min at room temperature. Supershifting antibodies were from Zymed.
Results
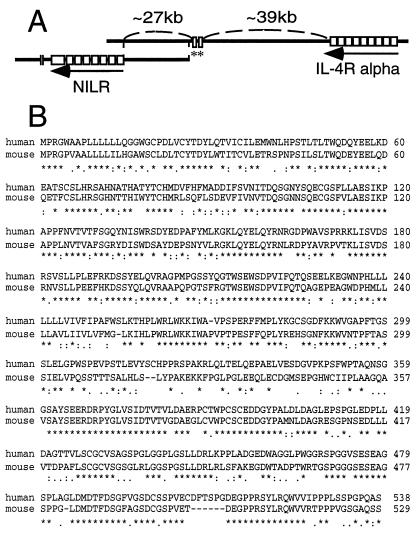
An ORF was identified on the genomic sequence of a bacterial artificial chromosome (BAC) clone (GenBank accession no. AC002303) (14) from chromosome 16p12 by using the gene prediction program genscan. The virtual gene was predicted to contain 9 coding exons comprising 568 amino acids, including a putative signal sequence, a transmembrane domain, four conserved Cys residues in the putative extracellular region, and a WSXWS motif, all of which suggested a type I cytokine receptor (2, 15, 16). It was provisionally named NILR. To clone NILR, we designed nested PCR primers 5′ and 3′ to the predicted start and stop codons, respectively. Nested PCR was performed with cDNA from bone marrow, fetal liver, leukocyte, placenta, spleen, and lung, and gel analysis revealed a strong band in all tissues except fetal liver. The bands were slightly smaller than the expected size; the smallest PCR product was from placenta. Several clones were sequenced. To evaluate possible PCR errors, the PCR product from bone marrow was labeled with 32P as a probe and used to screen a cDNA library prepared from mRNA of YT cells. Fifteen positive clones were confirmed by secondary and tertiary screening, two of which contained the full-length ORF. One of these cDNAs (clone 7) appeared to be a fusion cDNA that contained sequences from two other genes (human myeloid differentiation protein and ribosomal DNA). The other cDNA (clone 17) contained only the NILR sequence (Fig. 1). The deduced ORF is 538 amino acids long, the same as the longest clone from the PCR from bone marrow. The reason for the 30 amino acid difference from the virtual gene prediction is that a computer-predicted exon of 18 amino acids in the extracellular domain was not found in any cDNA clone, exon 3 was 26 amino acids shorter than predicted, and exon 5 was 14 amino acids longer than predicted (exons are numbered as in Table 1). NILR has four conserved Cys residues and a WSXWS motif, typical of type I cytokine receptors. Analysis with blast revealed that the most related protein is the human IL-2Rβ (17). The region between amino acids 116–350 of NILR is 27% identical to the corresponding region of IL-2Rβ. NILR contains the Box B1 region and conserved Trp typical of type I cytokine receptors in the membrane proximal region (18). As compared with the original BAC clone genomic DNA sequence, there were three differences in the NILR cDNA: nucleotide 1 in the 5′ untranslated region is a C versus an A in the BAC clone, nucleotide 1292 is an A in the cDNA versus a G in the BAC clone (thus codon 386 is an Arg in clone 17 but a Gly in the BAC clone), and nucleotide 1781 is a G in the 3′ untranslated region of the cDNA versus a C in the BAC clone. The other clone (clone 7) contained an ORF identical to that of the BAC clone. These differences could be polymorphisms or reverse transcriptase-generated differences. Interestingly, a second BAC clone (GenBank accession no. AC004525) (14) overlaps the first and contains two 5′ noncoding exons of NILR identified from the cDNAs and also contains exons encoding the IL-4 receptor α chain (Fig. 2A). The closest IL-4 receptor α chain gene (IL4RA) exons are only 39 kb 5′ to the NILR gene, in the same transcriptional orientation, suggesting that these genes are adjacent. The exon/intron structure of NILR is described in Table 1. Different cDNAs used either of two first exons (exon 1a or 1b) that spliced to exon 2 (which contains the ATG translation initiation site). In addition, PCR amplification reveals a transcript including sequences immediately 5′ to exon 2. This transcript is labeled as beginning with exon 1′, which includes exon 2 (Table 1). Whether this represents a third transcription initiation site or alternative splicing from exon 1a or 1b to a site upstream of exon 2 is unclear. Because the IL4RA and NILR genes are adjacent, it is conceivable that they arose by a gene duplication event. Like the NILR gene, the IL4RA gene has two noncoding exons, followed by three exons containing the signal peptide, and each of the two pairs of conserved Cys residues, respectively. After another exon are two exons containing the WSXWS motif and the transmembrane domain. Like most type I cytokine receptors, the NILR cytoplasmic domain is encoded by two exons, whereas the IL4R cytoplasmic domain is encoded by three exons. Thus, the overall organization of these genes is similar but not identical.
Table 1.
Genomic structure of NILR
| Exon | Exon length, bp | GenBank accession no.* | Location in genome† | Splice acceptor‡ | Splice donor‡ | Intron length, bp |
|---|---|---|---|---|---|---|
| 1a§ | 202 | AC004525 | 36742–36541 | Not applicable | ACAGAAGgtaattc | 27,425 |
| 1b§ | 120 | AC004525 | 36058–35939 | Not applicable | TCGGATGgtaagag | 26,823 |
| 1′¶ | 99 | AC002303/(AC004525) | 26245–26398/(9203–9050) | Not applicable | CAGGGAGgtaagtg | 4,226 |
| 2 | 65 | AC002303/(AC004525) | 26334–26398/(9115–9050) | cttgcagGCCCGTG | CAGGGAGgtaagtg | 4,226 |
| 3 | 103 | AC002303/(AC004525) | 30625–30727/(4823–4721) | cctccagGCTGGGG | TTACCTGgtaagta | 3,038 |
| 4 | 200 | AC002303/(AC004525) | 33766–33965/(1685–1484) | cttgaagGCAAGAC | GAGAGCAgtgagta | 5,274 |
| 5 | 155 | AC002303 | 39240–39394 | caccaagTCAAGCC | GGCTGTGgtgagga | 1,425 |
| 6 | 178 | AC002303 | 40820–40997 | ctggcagAGTCCGA | TCAGAGGgtaggtt | 457 |
| 7 | 100 | AC002303 | 41455–41554 | ttcccagAGTTAAA | TGTGGAGgtgaggc | 780 |
| 8 | 82 | AC002303 | 42285–42366 | ccctcagGCTATGG | CTTCAAGgtgagct | 730 |
| 9 | 1,540 | AC002303 | 44812–46351 | ctcacagAAATGGG | Not applicable |
AC004525 sequence is in reverse orientation with respect to the NILR transcript.
Intronic sequence is in lower case.
The 5′ boundaries of exons 1a and 1b are the 5′ end of cDNA clones and have not been proven to be the transcription initiation site.
Exon 1′ is identical to exon 2 except that it extends further 5′. Its 5′ boundary has not been mapped. See text.
Figure 2.
The NILR and IL4RA genes are adjacent, and human and murine NILR genes are highly related. (A) Schematic showing two overlapping BAC clones that together span the NILR gene. The BAC clone positioned to the left contains all NILR coding exons; the one on the right contains at least two noncoding exons of NILR (asterisks) and the IL4RA gene. Marked distances are approximate. (B) Alignment of deduced human and murine NILR amino acid sequences by using clustal w (11). Asterisks, colons, and periods indicate identical amino acids and conservative and semiconservative substitutions, respectively.
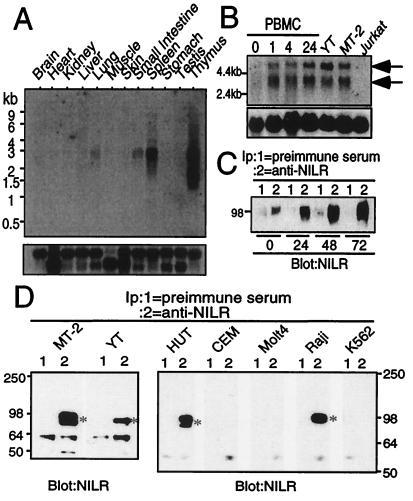
We next identified a full-length murine NILR cDNA by screening a murine thymus cDNA library with a human NILR probe. The deduced murine and human amino acid sequences are aligned in Fig. 2B. We did not identify cDNAs containing the first two coding exons, but 5′ rapid amplification of cDNA ends revealed that the mouse NILR amino acid sequence is 529 amino acids long. Human and murine NILR are 72% identical at the DNA level and 62% identical at the amino acid level. Extracellular Cys residues and WSXWS motif are conserved. Human and mouse NILR cytoplasmic domains both contain a Box B1 motif and six Tyr residues. To evaluate the tissue distribution of NILR, we performed Northern blot analyses using a mouse multitissue poly(A)+ RNA membrane. Expression was greatest in lymphoid tissue such as thymus and spleen (Fig. 3A). The principal transcript was 2.5–3.2 kb; thymus had a second band of 1.5–1.8 kb. A longer exposure suggested low-level NILR expression in lung and small intestine, possibly explained by lymphocytes in these tissues. Moreover, NILR is also expressed in bone marrow, given that we were able to PCR-amplify NILR from human bone marrow cDNA. Interestingly, in human PBMC, expression of NILR mRNA was low but induced after stimulation of the T cell antigen receptor with PHA within 1 h (Fig. 3B). This was confirmed at the protein level (Fig. 3C). Thus, like IL-2Rβ, NILR expression is increased after T cell stimulation with PHA (17).
Figure 3.
NILR mRNA and protein expression. (A) (Upper). NILR mRNA expression distribution in mouse tissue. (Lower) Hybridization with a control human β-actin probe. (B) NILR mRNA expression is induced by PHA in human PBMC. Cells were stimulated with PHA for the indicated times and poly(A)+ RNA was isolated. NILR mRNA expression was also evaluated in YT (a positive control because YT cells were the source of the cDNA library), MT-2, and Jurkat T cell lines. Arrows indicate NILR mRNA. (Lower) Hybridization with the pHe7 probe as a control (12). (C) NILR protein is induced by PHA in human PBMC. Cells were stimulated with PHA for the indicated time periods, and preimmune serum (lane 1) or anti-NILR antiserum (lane 2) immunoprecipitates were blotted with anti-NILR antiserum. For each immunoprecipitation, 3.0 × 107 cells were used. (D) NILR protein is expressed in MT-2, YT, HUT-102B2, and Raji cells, but not in CEM, Molt4, or K562 cells. Immunoprecipitates of preimmune serum (lane 1) or anti-NILR antiserum (lane 2) were blotted with anti-NILR antiserum. For each immunoprecipitation, 2.0–2.5 × 107 cells were used.
Immunoprecipitation and Western blotting with preimmune and anti-NILR antiserum also revealed the expression of this receptor on HTLV-I-transformed T cell lines (MT-2 and HUT-102B2), an NK-like cell line (YT), and a B cell line (Raji), but not on two T cell lines (Molt4 and CEM), or on a myeloid cell line (K562). These data suggest that activated T cells, B cells, and NK cells can express NILR. Western blotting revealed that NILR protein is ≈80–100 kDa, with some variation depending on the cell line (Fig. 3D), whereas the calculated molecular mass is 60 kDa, consistent with NILR being a glycoprotein. Indeed, there are five Asn-Xaa-Ser/Thr N-linked glycosylation sites and a number of potential O-linked glycosylation sites in the NILR extracellular domain.
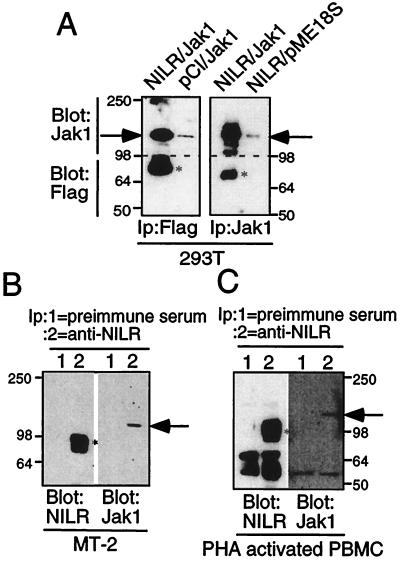
We next evaluated whether NILR could associate with Jak kinases, as is typical of type I cytokine receptors. Indeed, the Box B1 region that serves this function is present in NILR (19–22). Because of its similarity to IL-2Rβ, which interacts primarily with Jak1 (1), we first investigated the possible NILR–Jak1 interaction by using a 293T cell overexpression system in which FLAG-tagged NILR and Jak1 expression vectors were transfected. Jak1 was readily detected in anti-FLAG immunoprecipitates, and conversely, FLAG-tagged NILR was detected in anti-Jak1 immunoprecipitates (Fig. 4A). We confirmed this NILR–Jak1 association in both MT-2 cells and PHA-activated PBMC by demonstrating that anti-NILR antiserum was capable of coimmunoprecipitating Jak1 (Fig. 4 B and C). The Jak1–NILR association supports a possible role of NILR as an important signaling molecule, although the ligand remains unknown. Because of the homology of NILR to IL-2Rβ, we tested whether some of the cytokines related to IL-2 could bind to NILR in affinity-labeling experiments. Specifically, 125I-labeled IL-2, IL-4, IL-7, and IL-15 each bound to their known receptors, but we could not demonstrate binding of any of these cytokines to NILR alone or when NILR was coexpressed with γc (data not shown), suggesting that the NILR ligand may be a novel cytokine.
Figure 4.
Association of Jak1 with NILR. (A) FLAG-tagged NILR and Jak1 associate in transfected 293T cells. Indicated plasmids were transiently transfected. Samples were immunoprecipitated with anti-FLAG (Left) or anti-Jak1 (Right). The upper part of the membrane (>98 kDa) was blotted with anti-Jak1 and the lower part (<98 kDa) was blotted with anti-FLAG. Bands corresponding to NILR and Jak1 are indicated by asterisks and arrows, respectively. (B and C) Anti-NILR immunoprecipitates from HTVL-I-transformed MT-2 cells (B) and PHA-activated PBMC (C) contain Jak1. Preimmune serum (lane 1) or anti-NILR (lane 2) immunoprecipitates were divided; one part was blotted with anti-NILR antiserum (Left) and the other one was blotted with anti-Jak1 (Right). The asterisk and arrow indicate NILR and Jak1, respectively.
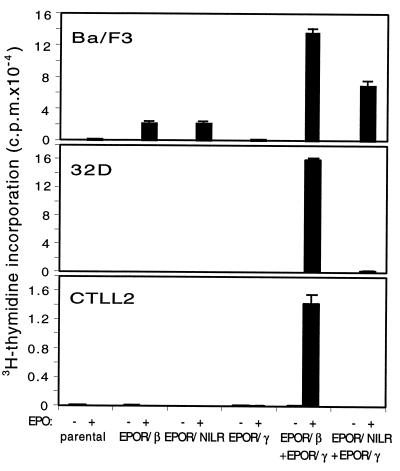
To further evaluate the signaling potential of NILR, we made chimeric EPOR/NILR, EPOR/IL-2Rβ, and EPOR/γc receptor retroviral vectors (see Materials and Methods). These were transduced into IL-3-dependent Ba/F3 or 32D cells or IL-2-dependent CTLL-2 cells. In 32D and CTLL-2, only cells expressing both EPOR/IL-2Rβ and EPOR/γc proliferated in response to EPO, and cells expressing EPOR/NILR did not proliferate even when EPOR/γc was coexpressed (Fig. 5). Interestingly, the EPOR/NILR chimera can mediate proliferation in the presence of EPO in Ba/F3 (but not 32D and CTLL-2 cells) (Fig. 5). Ba/F3 cells are also permissive to proliferation by the isolated EPOR/IL-2Rβ chimera as reported (23). Thus, analogous to IL-2Rβ, homodimerization of NILR can trigger proliferation in Ba/F3 cells, but unlike IL-2Rβ, heterodimerization of NILR with γc is not sufficient for potent proliferation in 32D or CTLL-2 cells, although a very low level of proliferation was seen in 32D cells (Fig. 5). It is conceivable that γc might contribute to NILR-mediated proliferation, given that higher proliferation was seen with EPOR/NILR + EPOR/γc than with EPOR/NILR alone in Ba/F3 cells. This difference was observed in two experiments where proliferation was evaluated at day 4 but not in single experiments where proliferation was evaluated at days 2 and 3. Identification of the NILR ligand will help to clarify the composition of the functional receptor and whether γc is a component of the receptor. Interestingly, Ba/F3 cells transduced with EPOR/NILR could be maintained long term in the presence of low-dose EPO (0.5 unit/ml) instead of IL-3.
Figure 5.
The NILR cytoplasmic domain can transduce a proliferative signal in Ba/F3 but not 32D and CTLL-2 cells. Ba/F3, 32D, and CTLL-2 cells expressing the indicated chimeric receptors were incubated with medium (open bars) or EPO at 100 units/ml (filled bars). After 92 h, 1 μCi of [3H]thymidine was added and cells were harvested 4 h later. As expected, Ba/F3 and 32D transfectants proliferated well in response to WEHI-3B-conditioned medium and CTLL-2 transfectants proliferated well in response to IL-2 (data not shown).
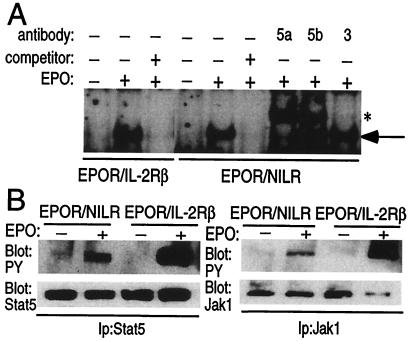
We next performed electrophoretic mobility-shift assays to investigate whether STAT proteins are activated by EPOR/NILR homodimerization (Fig. 6A). Similar-mobility DNA–protein complexes were induced by EPO in cells expressing either EPOR/IL-2Rβ or EPOR/NILR chimeras. Because IL-2Rβ mediates Stat5 activation (1), we evaluated the ability of anti-Stat5a, Stat5b, and Stat3 to supershift the EPOR/NILR-activated complex, and found that it contained Stat5a and Stat5b but not Stat3 (Fig. 6A). Stat5 tyrosine phosphorylation was also detected (Fig. 6B). Thus, like IL-2Rβ, NILR mediates the activation of Stat5, suggesting that the cytoplasmic domain of NILR may have Stat5-docking sites. Consistent with the association of Jak1 with NILR (Fig. 4), EPO could induce tyrosine phosphorylation of Jak1 in cells expressing the EPOR/NILR chimera, suggesting that Jak1 is a mediator of NILR signaling (Fig. 6B).
Figure 6.
Electrophoretic mobility-shift assays and anti-phosphotyrosine immunoblotting of Ba/F3 cells expressing the indicated chimeric receptor. (A) Cells maintained in EPO (0.5 units/ml) were deprived of growth factor and then stimulated or not stimulated with EPO (20 units/ml, 30 min). Nuclear extracts were then prepared and binding assays were performed. The arrow indicates a DNA-binding protein corresponding to Stat5 and the asterisk indicates the bands that are detected after supershifting with antibodies to either murine Stat5a or Stat5b. (B) Cells were deprived of growth factor and then stimulated or not stimulated with EPO (20 units/ml, 10 min). Stat5 and Jak1 immunoprecipitates were blotted with anti-Stat5, anti-Jak1, or anti-phosphotyrosine antibodies as indicated.
Discussion
In this study, we report the cloning of a type I cytokine receptor, denoted NILR, whose expression appears to be relatively restricted to lymphohematopoietic tissue. The pattern of expression in cell lines suggests that activated T cells, B cells, and NK cells can express the receptor. Whereas NILR was expressed in HTLV-I-transformed T cells and PHA-stimulated blasts, it was not detected in the Jurkat, CEM, and Molt4 T cell lines, consistent with expression primarily on highly activated T cells. It will be interesting to determine the potential role of the NILR ligand in B cell and NK cell biology and in the growth/survival of activated T cells and HTLV-I-mediated leukemogenesis.
Because NILR is most related to IL-2Rβ, we investigated whether NILR might represent an alternate receptor for IL-2 or related cytokines. However, the results were negative, suggesting that NILR is a receptor for a novel cytokine. Analogous to IL-2Rβ, an NILR homodimer was sufficient for growth of Ba/F3 but not 32D and CTLL-2 cells, indicating its signaling potential, but also indicating that at least one other chain is required for NILR-dependent signaling. It will be of great interest to identify the NILR ligand and the full composition of the NILR receptor.
NILR associates with Jak1, which is tyrosine phosphorylated after homodimerization of NILR, whereas preliminary studies provide no evidence for tyrosine phosphorylation of Jak2, Jak3, or Tyk2 under the same conditions. Electrophoretic mobility-shift assays revealed that Stat5 is activated after dimerization of the NILR cytoplasmic domain, which contains six Tyr, one or more of which might potentially be a docking site for Stat5.
Given the lymphohematopoietic-restricted expression, T cell antigen receptor inducibility, similarity to IL-2Rβ, and genetic linkage to IL-4 receptor α chain, NILR is a potentially very interesting and important molecule. The critical goals are to identify a ligand and disrupt the locus in mice as two means of clarifying NILR function. Accordingly, these are both areas of active investigation.
Acknowledgments
We thank Dr. Harvey Lodish for providing the murine EPOR cDNA and recombinant EPO.
Abbreviations
- NILR
novel interleukin receptor
- IL-2Rβ
IL-2 receptor β chain
- γc
cytokine receptor γ chain
- NK
natural killer
- PBMC
peripheral blood mononuclear cells
- HTLV-I
human T lymphotrophic virus type I
- PHA
phytohemagglutinin-L
- EPO
erythropoietin
- EPOR
EPO receptor
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
- BAC
bacterial artificial chromosome
Note Added in Proof.
NILR is also now being denoted as the IL-21 receptor (24).
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF269133 and AF269134).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200360997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200360997
References
- 1.Leonard W J, O'Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 2.Leonard W J. In: Fundamental Immunology. Paul W E, editor. Philadelphia: Lippincott; 1999. pp. 741–774. [Google Scholar]
- 3.Miyajima A, Mui A L, Ogorochi T, Sakamaki K. Blood. 1993;82:1960–1974. [PubMed] [Google Scholar]
- 4.Leonard W J, Shores E W, Love P E. Immunol Rev. 1995;148:97–114. doi: 10.1111/j.1600-065x.1995.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 5.Takeshita T, Asao H, Ohtani K, Ishii N, Kumaki S, Tanaka N, Munakata H, Nakamura M, Sugamura K. Science. 1992;257:379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- 6.Noguchi M, Yi H, Rosenblatt H M, Filipovich A H, Adelstein S, Modi W S, McBride O W, Leonard W J. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 7.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Landolt-Marticorena C, Williams K A, Deber C M, Reithmeier R A. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 11.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaczmarek L, Calabretta B, Kao H T, Heintz N, Nevins J, Baserga R. J Cell Biol. 1987;104:183–187. doi: 10.1083/jcb.104.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J X, Mietz J, Modi W S, John S, Leonard W J. J Biol Chem. 1996;271:10738–10744. [PubMed] [Google Scholar]
- 14.Loftus B J, Kim U J, Sneddon V P, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby M L, Barnstead M, Cronin L, et al. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- 15.Minami Y, Kitamura T, Miyazaki T, Taniguchi T. Annu Rev Immunol. 1993;11:245–268. doi: 10.1146/annurev.iy.11.040193.001333. [DOI] [PubMed] [Google Scholar]
- 16.Miyajima A, Kitamura T, Harada N, Yokota T, Arai K. Annu Rev Immunol. 1992;10:295–331. doi: 10.1146/annurev.iy.10.040192.001455. [DOI] [PubMed] [Google Scholar]
- 17.Hatakeyama M, Tsudo M, Minamoto S, Kono T, Doi T, Miyata T, Miyasaka M, Taniguchi T. Science. 1989;244:551–556. doi: 10.1126/science.2785715. [DOI] [PubMed] [Google Scholar]
- 18.Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, Taga T, Kishimoto T. Proc Natl Acad Sci USA. 1991;88:11349–11353. doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witthuhn B A, Quelle F W, Silvennoinen O, Yi T, Tang B, Miura O, Ihle J N. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 20.DaSilva L, Howard O M, Rui H, Kirken R A, Farrar W L. J Biol Chem. 1994;269:18267–18270. [PubMed] [Google Scholar]
- 21.Gurney A L, Wong S C, Henzel W J, de Sauvage F J. Proc Natl Acad Sci USA. 1995;92:5292–5296. doi: 10.1073/pnas.92.12.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner J W, Chen W, Young R L, Longmore G D, Shaw A S. J Biol Chem. 1995;270:6523–6530. doi: 10.1074/jbc.270.12.6523. [DOI] [PubMed] [Google Scholar]
- 23.Nelson B H, Lord J D, Greenberg P D. Nature (London) 1994;369:333–336. doi: 10.1038/369333a0. [DOI] [PubMed] [Google Scholar]
- 24.Parrish-Novak, J., Dillon, S. R., Nelson, A., Hammond, A., Sprecher, C., Gross, J. A., Johnston, J., Madden, K., Xu, W., West, J., et al. (2000) Nature (London), in press.