Abstract
The reported isolation of nanobacteria from human kidney stones raises the intriguing possibility that these microorganisms are etiological agents of pathological extraskeletal calcification [Kajander, E. O. & Çiftçioglu, N. (1998) Proc. Natl. Acad. Sci. USA 95, 8274–8279]. Nanobacteria were previously isolated from FBS after prolonged incubation in DMEM. These bacteria initiated biomineralization of the culture medium and were identified in calcified particles and biofilms by nucleic acid stains, 16S rDNA sequencing, electron microscopy, and the demonstration of a transferable biomineralization activity. We have now identified putative nanobacteria, not only from FBS, but also from human saliva and dental plaque after the incubation of 0.45-μm membrane-filtered samples in DMEM. Although biomineralization in our “cultures” was transferable to fresh DMEM, molecular examination of decalcified biofilms failed to detect nucleic acid or protein that would be expected from growth of a living entity. In addition, biomineralization was not inhibited by sodium azide. Furthermore, the 16S rDNA sequences previously ascribed to Nanobacterium sanguineum and Nanobacterium sp. were found to be indistinguishable from those of an environmental microorganism, Phyllobacterium mysinacearum, that has been previously detected as a contaminant in PCR. Thus, these data do not provide plausible support for the existence of a previously undiscovered bacterial genus. Instead, we provide evidence that biomineralization previously attributed to nanobacteria may be initiated by nonliving macromolecules and transferred on “subculture” by self-propagating microcrystalline apatite.
The identification of microorganisms, referred to by geologists as “nannobacteria”, has been inferred exclusively from the presence of coccoid-shaped particles, with diameters of approximately 0.1 μm, in scanning electron micrographs of rock and mineral surfaces (1). In addition, the observation of similar structures on the surface of a freshly fractured Martian meteorite has led to the question of whether nannobacteria are relics of primitive life (2). This possibility has, however, been challenged on the basis that such particles are too small to be free-living cells, at least according to contemporary definitions (3–5).
Possible biological evidence for the existence of a group of small microorganisms, collectively referred to by biologists as “nanobacteria”, has come from the reported cultivation of these “bacteria” from commercial lots of FBS, human serum, and also kidney and dental pulp stones (6–11). Nanobacteria were initially described as slow-growing, intracellular contaminants in cultured mammalian cells. They were subsequently propagated in the absence of host cells by prolonged incubation of FBS, but not γ-irradiated FBS (γ-FBS), in DMEM. Biomineralization of the culture medium is the hallmark of nanobacterial growth. Mineralized forms of nanobacteria have been identified grossly by reaction with Hoechst's DNA stain, von Kossa staining for calcification, electron microscopic examination, and the demonstration of a transferable nucleating activity for biomineralization (6). Based on results from 16S rDNA sequencing, bovine and human isolates of nanobacteria have been tentatively assigned to the α-2 subgroup of proteobacteria, which also includes Brucella and Bartonella species that occur within and outside of host cells (6, 8).
The reported identification of nanobacteria from serum, kidney, and dental pulp stones (6–11) and of nannobacteria on the tooth surface (12), raises important questions concerning the natural distribution of these unusual microorganisms, their occurrence as adventitious agents in biological products, and their association with pathological extraskeletal calcification (13). To address these questions, we initiated studies to isolate and characterize nanobacteria from FBS, human saliva, and dental plaque. Mineralized particles with all of the physical properties of nanobacteria (6), including their ability to initiate biomineralization on transfer to fresh DMEM, were successfully “cultured” from FBS and human oral samples. However, further molecular and microbiological characterization of such cultures has failed to provide any evidence of a living entity. Instead, our results suggest that biomineralization, like that previously attributed to nanobacteria (6), depends on the nucleating activities of nonliving biological macromolecules and of apatite itself.
Materials and Methods
Samples of FBS (Sigma), γ-FBS (FBS irradiated with 25–35 kGy; Sigma), whole unstimulated saliva that was freshly collected from 11 donors on various occasions, and dental plaque collected from 27 unidentified patients of the NIDCR Dental Clinic were cultured for nanobacteria by methods similar to those described (6). In most cases, 0.25 ml of FBS, γ-FBS, or saliva or the entire plaque sample from one individual were added to tubes containing 2 ml of DMEM (high glucose containing 4 mM l-glutamine, 1.8 mM CaCl2, and 0.9 mM NaH2PO4; BioWhittaker). In some cases, samples were added to DMEM containing 10% (vol/vol) γ-FBS to promote the possible growth of fastidious organisms. Tube contents were mixed and immediately filtered through 0.45-, 0.2-, or 0.1-μm sterile Millex filters (Millipore) into sterile, flat-bottom screw cap culture tubes (Nalge). Cultures were incubated horizontally at 37°C in 5% CO2/95% air for times of up to 2 months and examined periodically.
Biofilm material was scraped from DMEM-containing subcultures of saliva, pelleted by centrifugation, and washed with PBS to remove residual salivary components. Scanning electron microscopy (SEM) was performed by using a Hitachi S-3500N variable pressure scanning electron microscope equipped with a PGT-IMIX-PC EDS system. Transmission electron microscopy (TEM) was performed by using a JEOL 100-CXII for both negative staining and thin section observations. Biofilm material was embedded in Spurr's resin for thin sectioning. Biofilm-associated macromolecules were prepared by dialysis of washed biofilm material against changes of 0.5 M EDTA followed by PBS by using small Slide-A-Lyzer 10K Dialysis Cassettes (Pierce).
Platinum PCR SuperMIX (GIBCO/BRL), primers (Genosys, The Woodlands, TX), and Ultra Pure Water (Advanced Biotechnologies, Columbia, MD) were used for PCR, which was generally conducted for 30 or 35 cycles with annealing of primers for 40 sec at 55°C and extension for 1 min at 72°C. PCR products in control reactions lacking added template were consistently noted after 35 cycles of PCR with various primer pairs designed from the putative sequences of nanobacterial 16S rDNA (i.e., X98418 or X98419). Such products, which result from amplification of contaminating template, were detected even when all experimental manipulations were performed in a CleanSpot PCR/UV Workstation (Coy Laboratory Products, Ann Arbor, MI) with either freshly opened reagents from the manufacturers listed above or comparable reagents purchased from different manufacturers. PCR products were isolated after agarose gel electrophoresis by using a gel extraction kit (Qiagen, Chatsworth, CA) and sequenced with an ABI Automated DNA sequencer.
In control studies, DMEM, with or without 10% (vol/vol) γ-FBS, was incubated with selected phospholipids or apatite and examined periodically by SEM and energy-dispersive x-ray microanalysis (EDX) for evidence of mineralization. Phosphatidylinositol (Sigma) was added either in chloroform (14) or as an aqueous dispersion (15) to final concentrations ranging from 10 to 200 μg/ml in DMEM. Sodium azide was included with dispersed phospholipids at a final concentration of 0.1% in DMEM to maintain sterility. The procedure of Eanes et al. (16) was followed to prepare apatite in DMEM. Briefly, the concentrations of calcium and phosphate ions in DMEM were increased to 10 mM by the addition of sterile CaCl2 and NaHPO4 stock solutions adjusted to pH 7.4 with HCl. These additions resulted in rapid precipitation of amorphous calcium phosphate, which was incubated overnight to allow for the subsequent conversion to apatite (16). Five successive 1:10 transfers of the DMEM/apatite suspension to fresh DMEM were performed over a period of approximately 1 month (i.e., equivalent to the serial transfer of subcultures), resulting in continued formation of apatite from the concentrations of soluble calcium and phosphate ions present in DMEM.
Results and Discussion
Nanobacteria were previously cultured from greater than 80% of commercial lots of FBS surveyed but not from FBS after exposure to a high dose of γ-irradiation (6, 7). To verify these observations, we incubated samples of FBS and γ-FBS in separate flasks of DMEM under cell culture conditions and periodically examined these cultures for the appearance of nanobacteria. Our observations were remarkably similar to those of Kajander and Çiftçioglu (6). After incubation for approximately 3 weeks, microscopic particles that settled to the bottom of the culture vessel were noted in flasks of DMEM that contained 10% (vol/vol) FBS but not in those that contained 10% (vol/vol) γ-FBS. Moreover, the ability to form such particles was transferable by 1:10 dilution of FBS-containing cultures into flasks of fresh DMEM as described (6). Similar observations were made with DMEM cultures of membrane-filtered (0.22- or 0.45-μm) dental plaque extracts from 8 of 27 patients and saliva samples from each of 11 individuals. In fact, the apparent cultivation of nanobacteria was achieved more readily from human saliva than from FBS. Consistent with previous descriptions of nanobacterial growth (6), incubation periods for the initiation of biomineralization generally varied from a few days in serum-free cultures (i.e., saliva or plaque filtrate in DMEM alone) to a few weeks in γ-FBS-containing cultures [i.e., saliva or plaque filtrate in DMEM with 10% (vol/vol) γ-FBS]. The development of mineralized aggregates or biofilms was typically observed regardless of incubation conditions (Fig. 1). Six serial 1:10 transfers of selected cultures to fresh DMEM were successfully conducted over a 6-month period to demonstrate the life-like transferability of biomineralization.
Figure 1.
Mineralized biofilm formed in a subculture of 0.45-μm membrane-filtered human saliva in DMEM. Control culture medium without added saliva (left flask) and culture medium inoculated with a 3-week-old primary culture of membrane filtered saliva (right flask). Both flasks were incubated approximately 1 month under cell culture conditions and tipped upright before photography to reveal the biofilm (right flask).
SEM of biofilm material scraped from subcultures of membrane-filtered saliva, prepared after three or four serial transfers into fresh DMEM, revealed clusters of small coccoid particles as well as larger dome-shaped structures (Fig. 2a) similar to those previously identified as nanobacteria (6). Moreover, major peaks of calcium and phosphorus were detected by EDX of individual particles from our cultures, and the presence of microcrystalline apatite, similar to that found in human bone, was established by x-ray diffraction analysis of washed biofilm material (results not shown). TEM of thin sections (Fig. 2b) revealed coccoid cell-like structures, comparable to those previously identified as nanobacteria (6), within larger mineralized particles and forms earlier referred to as D shapes (7). Small coccoid-shaped particles (Fig. 2c) that resembled putative nanobacteria (6) also were observed by TEM of negatively stained biofilm material. Caution must be used in the identification of these particles as nanobacteria, however, because they also resemble typical inanimate structures previously observed in either amorphous calcium phosphate formed from sterile solutions of inorganic salts (16) or calcite formed in the presence of organic material (17).
Figure 2.
Electron and light microscopic images of nanobacteria-like particles scraped from DMEM-containing subcultures of 0.45-μm membrane-filtered saliva. (a) SEM of biofilm material. (Bar = 5 μm.) (b) TEM of nanobacteria-like particles observed in thin sections of biofilm material. (Bar = 1 μm.) (c) TEM of negatively stained biofilm material showing small coccoid-shaped particles. (Bar = 0.2 μm.) (d) Light micrograph of biofilm material stained with Hoechst 33258 showing nonspecific surface fluorescence of coccoid-shaped particles. (Bar = 1 μm.)
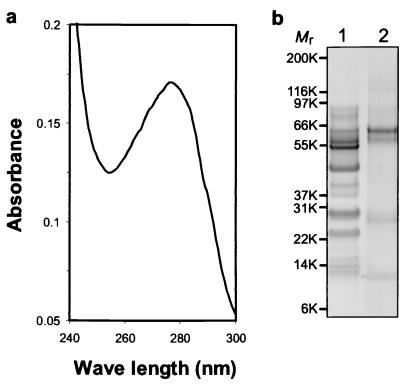
Staining of biofilm material with Hoechst 33258 at 5 μg/ml occurred as reported (6), but fluorescence was diffuse (Fig. 2d) and was not accepted as convincing evidence for the presence of nucleic acid. Nucleic acid also was not detected from samples of washed biofilm material (> 0.5 g) that were extracted with either 0.1% SDS or 1 M sodium phosphate (pH 7.5) or decalcified by dialysis against 0.5 M EDTA. Similarly, after decalcification and dialysis, no bacteria were pelleted (14,000 × g for 30 min), and the UV spectrum of the same preparation lacked an absorbance peak at 260 nm, typical of nucleic acid (Fig. 3a). Attempts to concentrate nucleic acid by ethanol precipitation from this and other soluble preparations of biofilm material were also unsuccessful; no material that stained with ethidium bromide was precipitated. Soluble protein was present in these dialyzed preparations as suggested by the absorbance peak near 280 nm in the UV spectrum (Fig. 3a), but the profile of this material revealed by SDS/PAGE (Fig. 3b) did not indicate a wide Mr range of polypeptides expected from a microbial proteosome. Rather, the electrophoretic pattern suggested the presence of a limited number of salivary components, presumably those that adsorb strongly to apatite.
Figure 3.
Biochemical examination of biofilm-associated macromolecules. Biofilm material from subcultures of 0.45-μm membrane-filtered saliva was washed with PBS and solubilized by dialysis against excess EDTA followed by PBS. (a) UV absorbance spectrum of the dialyzed preparation. (b) SDS/PAGE of the dialyzed preparation showing the Coomassie-stained profiles of membrane-filtered, whole human saliva (lane 1) and a comparable amount of biofilm-associated protein (lane 2). Mr of each molecular weight marker is indicated in thousands (K).
PCR, performed with primers designed from the 16S rDNA sequences reported for nanobacteria (EMBL X98418 and X98419; ref. 6), was used in further attempts to detect nanobacterial DNA. Amplification of a DNA fragment of the expected size (≈0.5 kilobases) was readily detected after 35 cycles of PCR, whether or not samples of putative nanobacteria cultured from saliva were decalcified by dialysis against EDTA followed by PBS. However, a PCR product of identical size was amplified to an equivalent extent in reaction controls performed without added template, thereby suggesting the detection of 16S rDNA from an environmental contaminant. This possibility was confirmed by the nucleotide sequence of the PCR product isolated from reaction controls, which was identical to the corresponding 16S rDNA sequence of Pseudomonas sp. (GenBank accession no. AF195876), but only 85.6% identical to the corresponding region of either putative nanobacterial sequence (Table 1).
Table 1.
Nucleotide sequence homology between 16S rDNA sequences
| 16S rDNA | bp | GenBank Accession No. | Percentage of identity*
|
||||
|---|---|---|---|---|---|---|---|
| AF195876 | D12789 | ATCC 43590 | X98419 | X98418 | |||
| PCR control† | 429 | — | 100 | 84.7 | 85.9 | 85.6 | 85.6 |
| N. sanguineum | 1,406 | X98418 | 82.5 | 99.0 | 99.6 | 98.6 | 100 |
| Nanobacterium sp. | 1,406 | X98419 | 82.7 | 97.8 | 99.6 | 100 | |
| ATCC 43590‡ | 515 | — | 86.1 | 98.8 | 100 | ||
| P. mysinacearum | 1,438 | D12789 | 82.5 | 100 | |||
| Pseudomonas sp. | 1,455 | AF195876 | 100 | ||||
Percentage of nucleotide sequence identity determined by bestfit (Genetics Computer Group, Madison, WI) alignment.
PCR product from control reactions (i.e., no added template) performed with primers designed from the reported sequences of nanobacterial 16S rDNA (i.e., X98418 or X98419). The forward primer, N1, was identical to the region from base 770 to 787, and the reverse primer, N2, complementary to the region from base 1,276 to 1,293 of each putative nanobacterial sequence. The sequence amplified was identical to that of AF195876 from position 868 to 1,296.
PCR product obtained from amplification of P. mysinacearum (ATCC 43590) DNA with primers N1 and N2. The sequence of the amplified product was identical to the 16S ribosomal DNA of P. mysinacearum HM35 (accession no. AJ011330) from position 785 to 1,299, except at positions 1,203 and 1,204, which were GC rather than CG.
Sequence comparisons revealed that the 16S rDNA sequences reported for nanobacteria are, however, highly homologous to the 16S rDNA of Phyllobacterium myrsinacearum, another microorganism identified as a source of contaminating 16S rDNA in PCR studies (18). Thus, as shown in Table 1, sequence X98418 of Nanobacterium sanguineum and X98419 of Nanobacterium sp., which are 98.6% identical to each other, are also 99.0% and 97.8% identical, respectively, to sequence D12789 of P. myrsinacearum (strain IAM 13584). As a control, we found that the sequence amplified from P. myrsinacearum ATCC 43590 with our primers (i.e., N1 and N2 defined in Table 1) was nearly identical to the putative nanobacterial sequences (i.e., X98418 and X98419) and also to that of P. myrsinacearum (i.e., D12789) deposited in GenBank in 1992. These findings assume heightened significance, because the reported PCR studies of Kajander and Çiftçioglu (6) do not include results from experimental controls or criteria for primer selection. In accordance with our interpretation, other investigators (19) also have recently suggested that the 16S rDNA sequences of nanobacteria could be PCR artifacts.
Although “culture properties” of nanobacteria have been described (6), credible bacteriological evidence of culture is sorely lacking, because individual cell growth and enumeration of nanobacteria have not been demonstrated. Putative nanobacteria have been detected with monoclonal antibodies prepared against mineralized preparations (8), but the immunochemical specificity of these reagents has not been established (8, 10). The appearance of mineralized nanobacterial colonies on modified Loeffler medium prepared in DMEM has also been described (6), but our attempts to cultivate or enumerate nanobacteria on standard Loeffler medium, in Ca2+-free DMEM, or in other bacteriological broths or agar media were unsuccessful (data not shown). Significantly, many reported properties of nanobacteria, including their resistance to several antibiotics and to high temperature (6), are based on the assumption that the initiation or transfer of biomineralization is an indicator of microbial growth. Similarly, the apparent sensitivity of nanobacteria to tetracycline and, especially, to citrate (10) may reflect the inhibitory effects of these compounds on calcification rather than on microbial growth. The use of biomineralization as an indicator of growth and the mineralized appearance of putative nanobacteria (cf. Fig. 2) have led to suggestions that nanobacteria occur within protective calcified envelopes or “dwellings” (6). In apposition with these interpretations, we found that biomineralization, although generally initiated or transferred by 1:10 dilutions of saliva or cultured biofilm material into fresh DMEM, respectively, was not initiated over an 8-week incubation period after 1:100 or 1:1,000 dilutions of these inocula. One might reasonably expect that bacterial regrowth after the latter dilutions would trigger biomineralization. In addition, we found that biomineralization was not only resistant to the presence of several antibiotics (100 μg/ml chloramphenicol, rifampin, kanamycin, or erythromycin) and heat (i.e., saliva/DMEM mixture heated at 90°C for 1 h before membrane filtration) as anticipated (20) but also unaffected by the continuous presence of easily diffusible 0.1% sodium azide which inhibits respiration. These observations not only question the association of biomineralization with bacterial growth but also, more importantly, rule out the presence of a respiration-dependent entity in putative nanobacterial cultures.
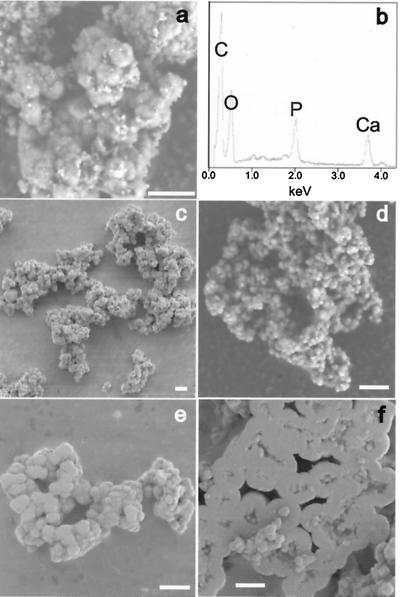
The Ca2+-nucleating activity detected previously in FBS was not only sensitive to a high dose of γ-irradiation (30 kGy) but was lost after 0.05-μm membrane filtration (6, 7). Similarly, we observed putative nanobacteria from FBS but not from γ-FBS and found that the nucleating activity of saliva was lost after 0.1-μm membrane filtration. Rather than affecting the viability or presence of nanobacteria, ionizing radiation (21) and membrane filtration may instead damage or remove nonliving macromolecular nucleators of biomineralization, for example, phospholipid complexes in serum (22) or lipid-protein complexes in saliva (23, 24). The nucleating activity of phospholipids (25, 26) was previously demonstrated by the formation of calcified spherules with diameters of approximately 0.2 μm from the interaction of phosphatidylinositol or phosphatidylserine with calcium and phosphate ions in a nonpolar environment (14). After incubation of phosphatidylinositol in DMEM for approximately 2 weeks, we also observed small, mineralized particles (Fig. 4a) and demonstrated the major presence of calcium and phosphate in such preparations by EDX (Fig. 4b). These results can be explained in terms of the relatively high concentrations of calcium and phosphate ions in DMEM (i.e., 1.8 mM and 0.9 mM, respectively), which are metastable and comparable to those used in studies of in vitro biomineralization (26).
Figure 4.
SEM of nanobacteria-like particles formed by incubation of phosphatidylinositol or apatite in DMEM. (a) Washed biofilm material formed by incubation of DMEM with 20 μg/ml phosphatidylinositol for approximately 2 weeks. (b) Detection of C, O, P, and Ca by EDX analysis of the sample shown in a (the sample was mounted on a carbon tab). (c–f) Washed apatite particles, formed initially from sterile solutions of CaCl2 and NaHPO4 in DMEM, after five 1:10 transfers into fresh DMEM. (Bars = 5 μm.)
Serum or saliva components that are potential nucleators of biomineralization in primary nanobacterial cultures would eventually be diluted out on subculture, and thus, these components may not account for the long-term transferability of biomineralization to fresh medium. However, apatite is also a nucleator (27). In fact, we found that this mineral, formed from sterile solutions of CaCl2 and NaHPO4 in DMEM, promoted further apatite formation, even after repeated dilution (i.e., 1:10) of the DMEM/apatite suspension to fresh DMEM. In addition, SEM of apatite formed under such conditions and, scraped from the culture flask, revealed clusters of small coccoid-shaped particles (Fig. 4 c–e) and dwelling-like structures (Fig. 4f) that closely resembled putative nanobacteria cultured either from saliva (Fig. 2a), FBS (6), or human kidney stones (9). Thus, the effect of apatite on metastable concentrations of calcium and phosphate ions in DMEM is sufficient to account for both the sustained, life-like transferability of biomineralization and the wide range of morphological forms previously attributed to nanobacteria (6).
Our studies of nanobacteria were initiated to confirm and extend the molecular and bacteriological properties of these unusual small microorganisms. We reasoned that, if nanobacteria are as widespread as suggested (refs. 6–11 and 20; i.e., present in 80% of commercial lots of FBS, most kidney stones, and dental pulp stones), then these microorganisms should exist in FBS available in the U.S. and in human oral samples. Indeed, putative nanobacteria were cultured from such samples by methods identical to those described by Kajander and Çiftçioglu (6). Further detailed examination of nanobacteria derived mainly from samples of human saliva revealed all of the expected properties, including the transferability of biomineralization. Definitive evidence in support of a living entity as the causative factor of biomineralization observed in such cultures was, however, not obtained from molecular and bacteriological analyses. Significantly, the 16S rDNA sequences ascribed singularly to nanobacteria (6, 8) were found to be virtually identical to sequences reported in 1992 for P. mysinacearum, thereby indicating that they were incorrectly assigned to a new genus by Kajander and Çiftçioglu (6, 8). The lack of other well defined biochemical, immunochemical, or molecular criteria for the identification of nanobacteria severely limited further comparisons of our findings with those of Kajander et al. (6–11, 20) and also emphasized a crucial issue. The potential existence of unusual microorganisms such as nanobacteria cannot be formally excluded based on negative evidence; rather, their existence must be established by a defining set of unambiguous criteria.
The accumulated evidence, including our interpretation that environmental contamination accounts for the putative nanobacterial 16S rDNA sequences reported (6), the limited number and ambiguous nature of the defining properties of nanobacteria, and the lack of bacteriologic evidence of culturability, all fail to provide reasonable support for the existence of small microorganisms referred to as nanobacteria. Moreover, our results indicate that biomineralization, like that attributed to the growth of nanobacteria in DMEM (6), can be triggered by biological macromolecules including phospholipids as described (14, 25, 26) and can be continued on dilution to fresh medium by apatite itself. These observations favor an alternative interpretation of nanobacteria-induced biomineralization (6) but do not diminish the importance of further efforts to define the underlying basis of pathological extraskeletal calcification. Indeed, continued studies to identify and characterize the primary nucleators of these important clinical conditions are needed, regardless of whether the molecules in question are of host and/or microbial origin.
Acknowledgments
We thank the staff of the National Institute of Dental and Craniofacial Research Dental Clinic for sample collection, William DiMichele, Jacob Donkersloot, Larry Fisher, Richard Kenney, and Michael Schmidt for constructive comments and manuscript review, and David Eanes for performing x-ray diffraction analysis and helpful discussions.
Abbreviations
- EDX
energy-dispersive x-ray microanalysis
- γ-FBS
γ-irradiated FBS
- SEM
scanning electron microscopy
- TEM
transmission electron microscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Folk R L. J Sediment Petrol. 1993;63:990–999. [Google Scholar]
- 2.McKay D S, Gibson E K, Jr, Thomas-Keprta K L, Vali H, Romanek C S, Clemett S J, Chillier X D F, Maechling C R, Zare R N. Science. 1996;273:924–930. doi: 10.1126/science.273.5277.924. [DOI] [PubMed] [Google Scholar]
- 3.Maniloff J. Science. 1997;276:1776. doi: 10.1126/science.276.5320.1773e. (lett.). [DOI] [PubMed] [Google Scholar]
- 4.Nealson K H. Science. 1997;276:1776. (lett.). [PubMed] [Google Scholar]
- 5.Psenner R, Loferer M. Science. 1997;276:1776–1777. (lett.). [PubMed] [Google Scholar]
- 6.Kajander E O, Çiftçioglu N. Proc Natl Acad Sci USA. 1998;95:8274–8279. doi: 10.1073/pnas.95.14.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Çiftçioglu N, Kuronen I, Xkerman K, Hiltunen E, Laukkanen J, Kajander E O. In: Vaccines 97. Brown F, Burton D, Doherty P, Mekalanos J, Norrby E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 99–103. [Google Scholar]
- 8.Çiftçioglu N, Kajander E O. Pathophysiology. 1998;4:259–270. [Google Scholar]
- 9.Çiftçioglu N, Björklund M, Kuorikoski K, Bergström K, Kajander E O. Kidney Int. 1999;56:1893–1898. doi: 10.1046/j.1523-1755.1999.00755.x. [DOI] [PubMed] [Google Scholar]
- 10.Hjelle J T, Miller-Hjelle M A, Poxton I R, Kajander E O, Ciftcioglu N, Jones M L, Caughey R C, Brown R, Millikin P D, Darras F S. Kidney Int. 2000;57:2360–2374. doi: 10.1046/j.1523-1755.2000.00096.x. [DOI] [PubMed] [Google Scholar]
- 11.Çiftçioglu N, Çiftçioglu V, Vali H, Turcott E, Kajander E O. SPIE Proc. 1998;3441:130–135. [Google Scholar]
- 12.Folk R L. Science. 1997;276:1777. (lett.). [Google Scholar]
- 13.Carson D A. Proc Natl Acad Sci USA. 1998;95:7846–7847. doi: 10.1073/pnas.95.14.7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore J M, Nichols G, Jr, Wuthier R E. Science. 1971;172:1339–1341. doi: 10.1126/science.172.3990.1339. [DOI] [PubMed] [Google Scholar]
- 15.Abramson M B, Katzman R, Gregor H P. J Biol Chem. 1964;239:70–76. [PubMed] [Google Scholar]
- 16.Eanes E D, Termine J D, Nylen M U. Calcif Tissue Res. 1973;12:143–158. doi: 10.1007/BF02013730. [DOI] [PubMed] [Google Scholar]
- 17.Kirkland B L, Lynch F L, Rahnis M A, Folk R L, Molineux I J, McLean R J C. Geology. 1999;27:347–350. [Google Scholar]
- 18.Tanner M A, Goebel B M, Dojka M A, Pace N R. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitcher D G, Fry N K. J Infect Dis. 2000;40:116–120. doi: 10.1053/jinf.2000.0635. [DOI] [PubMed] [Google Scholar]
- 20.Bjorklund M, Çiftçioglu N, Kajander E O. SPIE Proc. 1998;3441:123–129. [Google Scholar]
- 21.Edwards J C, Chapman D, Cramp W A, Yatvin M B. Prog Biophys Mol Biol. 1984;43:71–93. doi: 10.1016/0079-6107(84)90004-x. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg M, Lecolle S, Vermelin L, Benghezal A, Septier D, Godeau G. Calcif Tissue Int. 1999;65:66–72. doi: 10.1007/s002239900659. [DOI] [PubMed] [Google Scholar]
- 23.Larsson B, Olivecrona G, Ericson T. Arch Oral Biol. 1996;41:105–110. doi: 10.1016/0003-9969(95)00077-1. [DOI] [PubMed] [Google Scholar]
- 24.Rølla G, Rykke M, Gaare D. Adv Dent Res. 1995;9:403–409. [Google Scholar]
- 25.Vogel J J, Boyan-Salyers B D. Clin Orthop Relat Res. 1976;118:230–241. [PubMed] [Google Scholar]
- 26.Goldberg M, Boskey A L. Progr Histochem Cytochem. 1996;31:1–187. doi: 10.1016/s0079-6336(96)80011-8. [DOI] [PubMed] [Google Scholar]
- 27.Boskey A L. J Cell Biochem. 1998;30–31:83–91. [PubMed] [Google Scholar]