Abstract
The methanarchaeon, Methanococcus jannaschii, a hyperthermophilic, autotrophic, and strictly hydrogenotrophic inhabitant of submarine hydrothermal vents, was cultivated in a reactor at two hydrogen partial pressure (pH2) values, 178 kPa (high) and 650 Pa (ultralow), and the cells were subjected to a comparative proteome analysis. From these studies, it was discovered that, when pH2 was high and the cell density was low (a combination representing a hydrogen-excess condition), the cells possessed very low or undetectable levels of four flagella-related polypeptides (FlaB2, FlaB3, FlaD, and FlaE); electron microscopic examination showed that most of these cells were devoid of flagella. Flagella synthesis occurred when hydrogen became limiting either at high cell density under high pH2 or at low cell density under low pH2. The results from a pH2-shift experiment corroborated the above observations. The pH2-dependent changes in the levels of two methanogenic enzymes (MTD and HMDX) were as expected, and thus they served as internal controls. To our knowledge, this is the first example for the regulation of expression of flagella by hydrogen in any domain of life and for a control of any kind on flagella synthesis in the archaea. Our work also provides the only known example for each of the following: (i) the pure culture cultivation of a methanogen at an ultralow, near ecologically relevant pH2; (ii) experimental functional genomics for M. jannaschii; and (iii) the use of proteomics with M. jannaschii.
Methanococcus jannaschii is an autotrophic, strictly hydrogenotrophic, and hyperthermophilic (optimal growth temperature, 85°C) methanarchaeon isolated from material collected from the base of a “white smoker” submarine hydrothermal vent (1). It is also the first archaeon for which the sequence of the entire genome was determined (2). The first analysis of these data brought many surprises, of which was paramount the realization that only 38% of the predicted protein-coding regions could be assigned even a putative cellular function with confidence (2). This conclusion remains largely valid, even after several investigators applied various algorithms to decipher the functions for most of the ≈1.7-megabase DNA sequence data with no intelligible biology (3). A direct analysis of M. jannaschii remained mostly stalled because of investigators' inability to manipulate the genome genetically or to perform subcellular biochemistry experiments that required significant cell mass. Recently, we have removed the latter hurdle by developing a protocol for mass culturing the organism to high cell densities (4). More importantly, our protocol allowed for the first time a study of the behavior of this archaeon under a variety of highly defined and reproducible conditions and at very low cell densities; the last attribute is especially important, because it might simulate the natural habitat for M. jannaschii, in the hydrothermal vents. We have decided to take advantage of this opportunity to study the response of M. jannaschii to a variety of stimuli. We have used the 2-D gel electrophoresis-based proteome analysis method for monitoring intracellular activities. M. jannaschii is a strict hydrogenotroph, and hydrogen is present in vent-effluent waters (1, 5). Because oxygen is absent inside these vents (channels and reservoirs included; ref. 5), HS− + O2-based energy metabolism is not possible; hydrogen may act as one of the major primary energy sources that sustains the living world in hydrothermal vents, making M. jannaschii one of the primary producers. For these reasons, we have studied the effect of pH2 on the behavior of this organism. The results presented in this communication clearly show that a combination of reactor culture experiments and proteomics can be of great help in discovering behavioral aspects of M. jannaschii that are not always logically conceivable.
Experimental Procedures
Growth Experiments.
M. jannaschii JAL-1 (1) was obtained from David R. Boone (Oregon Collection of Methanogens, Portland, OR). All growth experiments were conducted in an instrumented stainless steel constantly stirred tank reactor (12-liter working volume) as described previously (4), and the specific details were as follows. Medium 1 (4) with 50 μM Se was used. The impeller tip speed was 235.5 cm−1, and the vessel was maintained at a positive pressure of 1.24 × 105 Pa. The gases were supplied at the following rates (ml⋅min−1; as measured at a pressure of 1 atm or 1.01 × 105 Pa): CO2, 4,800; H2S (a mixture of N2 and H2S, 90:20 vol/vol), 215; H2, 19,200 for a pH2 of 178 kPa and 70 for a pH2 of 0.65 kPa; N2, 19,130 (only for the low pH2 culture, as a volume make-up gas). Because all experiments were conducted at low cell densities, pH of the culture remained unperturbed and no base addition occurred. For the same reason, turbidity values did not truly reflect the progress of a culture. Thus, the cell density was followed by cell counts, which were determined by use of a Petroff Hauser Bacteria Counter (Hauser Scientific, Blue bell, PA) and a Model BH-2 microscope (Olympus, Tokyo, Japan) fitted with an epifluorescence attachment. A culture sample for proteome analysis was collected under N2–CO2 (80:20, vol/vol) by maintaining strict anaerobic conditions. A sealed and anaerobic 530-ml serum bottle (4) filled with N2–CO2 to 0.7 × 104 Pa overpressure was used as the sample collection vessel. The sample was cooled rapidly (within 10–15 seconds) in an ice–water bath as it was being collected.
Electron Microscopy.
The cells from 1–2 ml of a cooled culture sample were pelleted aerobically via centrifugation at 9000 × g for 2 min at 4°C and then were resuspended in 200 μl of modified Karnovsky's fixative [0.1 M K-cacodylate buffer (pH 7.2)/2.5% paraformadehyde/2% gluteraldehyde (6)] and stored at 4°C until used. The cells were negatively stained with ammonium molybdate [0.7 g⋅liter−1 (pH 6.2) (7)], because, in our hands, it helped to visualize the flagella better than uranyl acetate. The stained cells were examined and photographed by use of a Hitachi (Tokyo) H600 transmission electron microscope at the Center for Microscopic Imaging, College of Veterinary Medicine, University of Illinois.
Preparation of Cell Extracts and 2-D Gel Electrophoresis.
From a cooled culture sample (≈500 ml), the cells were harvested aerobically at 4°C in two steps: centrifugation at 10,500 × g for 30 min, resuspension of the first stage pellet in the entrained supernatant, and recentrifugation at 12,000 × g for 10 min. The final pellet was stored at −20°C. The target of our analyses was the whole cell extract (cytosolic and membrane fractions), and it was prepared as follows. The frozen cell pellets were resuspended in ice-cold 25 mM potassium phosphate buffer (pH 7), containing Complete Protease Inhibitor mixture (1 tablet per 50 ml) (cat. no. 1697 498; Roche Molecular Biochemicals). This suspension was passed three times through an ice-chilled French pressure cell at 1.25 × 108 Pa. The resulting cell lysate was centrifuged in a microcentrifuge at 9,000 × g for 5 min. The pellet from this step was discarded, and the supernatant was saved at −20°C until used.
The general method for the 2-D gel electrophoresis was that of O'Farrell (8), and the separations were performed by Kendrick Laboratories (Madison, WI). Each frozen cell extract was thawed and diluted to a protein concentration of 1 μg/μl with a buffer containing 60 mM Tris⋅HCl (pH 6.8), 5% SDS, 5% 2-mercaptoethanol, and 10% glycerol. This protein solution was boiled for 5 min, and then a 50-μl portion of it was subjected to isoelectric focusing (IEF) by use of a 2.3-mm (diameter) × 16-cm gel containing 2% BDH (pH 4–8) ampholines for 9,600 V-h. A 10% acrylamide slab gel (13 cm × 14.5 cm × 0.75 cm) was used for the second dimension denaturing gel electrophoresis. The fixed slab gels were stained appropriately and dried between sheets of cellophane; for the purpose of visual comparative analyses, the gels were stained with silver, but when a gel served as a source of peptide samples for identity determination (see below), it was stained with either Coomassie blue or special silver (9).
In-Gel Digestion, Mass Spectrometry, and Data Analysis.
Each polypeptide spot of interest was subjected to proteolysis by use of the in-gel digestion technique (10, 11) employing either trypsin or Lys-C (Roche Molecular Biochemicals). The resultant peptides were extracted from the gel piece with 50% acetonitrile in 2% trifluoroacetic acid. The extract was dried, and the dry material was resuspended in a matrix solution (10 mg/ml 4-hydroxy-α-cyanocinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid) that also contained two internal standards, angiotensin and bovine insulin. This final solution was used in matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometric analysis by use of a PerSeptive Voyager DE-RP mass spectrometer (PerSeptive Biosystems, Framingham, MA) in the linear mode. Both the in-gel digestion and mass spectrometric analysis of the digest were performed in the Protein Chemistry Core Facility of the Howard Hughes Medical Institute at Columbia University. The programs ms-fit and profound (P. R. Baker and K. R. Clauser, http://prospector.ucsf.edu; http://prowl.rockefeller.edu/cgi-bin/ProFound) were used to search the NCBI nonredundant (nr) database; here, the measured mass values for the proteolytic fragments were the inputs. The primary structure of each candidate polypeptide identified from the database searches was analyzed for corresponding theoretical fragments by use of the peptidemass program (http://www.expasy.ch/tools/peptide-mass.html), and the masses of these theoretical fragments were compared with the experimental data (Table 1). An identity assignment for a polypeptide spot was considered valid if at least 25% of the identified polypeptide's primary structure was represented by the fragment ions appearing in the corresponding mass spectrum (12).
Table 1.
Identities for selected polypeptide spots from 2-D gels for M. jannaschii cell extracts
| Spot number* (mass, kDa)* (digesting enzyme) | Polypeptide identified (Calculated mass, Da/pI), Accession number | MALDI mass† (MH+, average), Da | The peptide sequence consistent with observed mass‡ (start-end position in the parent polypeptide) {calculated average size, Da} | Δ, Da (observed mass −calculated mass) | Coverage§ [pro-peptide region excluded from calculations]¶ |
|---|---|---|---|---|---|
| LH178 (30 kDa) (Trypsin) | MJ0892/FlaB2 (21,155/4.73) Q58302 | 3681.31 2887.26 2327.35 | TTVTGSVIPEFGAPAVIEFTTPAAYLSTQEVIQLQ (183–217) {3681.174} AMATGKESTEQVASGLSTLQVIGIHDNK (47–74) {2887.234} ESTEQVASGLSTLQVIGIHD NK (53–74) {2327.553} | 0.136 0.026 −0.203 | 29% [pro-peptide, residues 1–12] |
| LH179 (29 kDa) (Trypsin) | MJ0893/FlaB3 (21,720/4.86) Q58303 | 2935.11 2283.28 1998.02 1743.88 1416.52 | LAILVSPNVGDEIDLSSTIVTISNGDYK (79–106) {2935.298} ERIYGEVIPEFGASGIIEFR (184–203) {2283.588} IYGEVIPEFGASGIIEFR (183–203) {1998.285} AIIAINVGDVFGGIMPR (167–183) {1744.105} APSTFSEHVVTLQ (204–216) {1416.573} | −0.188 −0.308 −0.568 −0.225 −0.053 | 37% [pro-peptide, residues 1–11] |
| LH174 (16 kDa) (Trypsin) | MJ0895/FlaD (39,950/5.91) Q58305 | 2198.28 1958.09 1714.84 1405.55 1152.35 | LAGRPIDSEILEMLEIEIR (312–330) {2198.582} DKLSPSDHIVSLLYIEK (295–311) {1958.262} LSPSDHIVSLLYIEK (297–311) {1714.999} ITFDEEELRPR (284–294) {1405.550} ITFDEEELR (284–292) {1152.246} | −0.302 −0.172 −0.159 0.0 0.104 | 14% (or 27% for a C-terminal ∼16 kDa fragment; 207–342 stretch; pI, 5.14) |
| LH175 | MJ0896/FlaE | 1797.94 | LNITDHIISLLFIER (95–109) {1798.135} | −0.195 | 27% |
| (15 kDa) | (15,892/5.02) | 1715.84 | ISAELLDKIEWELR (115–128) {1715.987} | −0.147 | |
| (Trypsin) | Q58306 | 1013.06 | KGAEQFYGI (132–140) {1013.138} | −0.078 | |
| LH176 | MJ1035/MTD | 2483.90 | MVEEVKPDFIIYIGPNPAAPGPK (55–77) {2483.928} | −0.028 | 35% |
| (30 kDa) | (30,306/5.86) | 2234.64 | AREILSQSGIPAVIIGDAPGLR (79–100) {2234.603} | 0.037 | |
| (Trypsin) | Q58441 | 2136.37 | REFLDPVEMALFNADVIR (125–142) {2136.473} | −0.103 | |
| 2007.29 | EILSQSGIPAVIIGDAPGLR (81–100) {2007.337} | −0.047 | |||
| 1980.08 | EFLDPVEMALFNADVIR (126–142) {1980.286} | −0.206 | |||
| 1958.09 | EAEKYIPIVASAHEMIR (220–236) {1958.283} | −0.193 | |||
| 1500.73 | YIPIVASAHEMIR (1500.799) {224–236} | −0.069 | |||
| 1471.65 | AVEAMEFTNPYAK (182–194) {1471.668} | −0.018 | |||
| 1053.19 | AMAAFTIAEK (197–206) {1053.261} | −0.071 | |||
| HH023 | MJ0715/HMDX | 2477.83 | QILISLQTIASIIETSGMEGLMK (236–258) {2477.977} | −0.147 | 42% |
| (37 kDa) | (36,854/5.69) | 2384.55 | AGHDVVLSEPNRDVMSDDLWK (42–62) {2384.625} | −0.075 | |
| (Lys-C) | Q58125 | 2208.3 | RRDVGISSMHPTGVPGTPSQK (135–155) {2208.500} | −0.2 | |
| 1967.14 | ISIYGAGNQRLYLEQLK (3–19) {1967.275} | −0.135 | |||
| 1916.08 | NMLIDERQEDLNLALK (271–286) {1916.199} | −0.119 | |||
| 1800.03 | FGGEPPYGGAGMAIEFAK (24–41) {1800.038} | −0.008 | |||
| 1660.86 | DISQTYLVAPQALIK (300–314) {1660.951} | −0.091 | |||
| 1435.59 | SAVEGMIRRSSNK (323–335) {1435.641} | −0.051 |
See Fig. 1 C–J for the location of a spot in the gels; the apparent molecular mass was derived from the migration distances in these gels.
Masses not matched: LH178 (4153.87, 4111.14, 3855.81, 3719.01, 3270.14, 3313.42, 3270.14, 2954.86, 2674.74, 2082.78); LH179 (4095.09, 3680.52, 3556.75, 3395.36, 1759.52, 1416.52); LH174 (2214.39, 1754.47, 1737.81, 1441.46, 1425.48, 1225.76); LH175 (2551.73, 2198.11, 1844.01, 1405.52); LH176 (1861.52, 1516.34, 1483.84, 1068.88); HH023 (2400.67, 2322.55, 2288.5, 2225.72, 1989.44, 1932.11, 1821.8, 1682.87, 1319.47).
Matched by use of the ProFound and the MS-Fit programs. The general search parameters were: MW of the intact polypeptide, a range based on the second dimension electrophoresis data (or a higher value, if the first search did not yield a good match; this could happen due to cleavage or processing of the full-length polypeptide or due to an error in assigning an ORF length or a start site in the databases); digest chemistry or type, trypsine or Lys-C (as applicable); cysteine modification, by acrylamide; maximum number of missed cleavages, 1 for trypsin and 0 for Lys-C; peptide mass type, average. The program-specific parameters were as follows. For ProFound: taxonomic category, archaebacteria; search for, single protein only; number of top candidate proteins, 10; peptide masses, M + H+; mass tolerances, 0.5 Da. 2. For the MS-Fit program: species, Methanococcus jannaschii; protein pI, 3–10; N terminus, hydrogen; C terminus, free acid; maximum reported hits, 5; search mode, identity; minimum number of matches with no amino acid substitutions, 1; peptide mass shift, 40.1 Da; possible modifications mode, default; user defined modification 1, phosphorylation of S, T, Y; minimum number of peptides required to match; 4 (or variable); Pfactor, 0.4; peptide mass tolerance, 0.5 Da.
The portion of the identified polypeptide's amino acid sequence covered by the matched fragments.
The propeptide sequence was located by comparison with the corresponding M. voltae homolog, for which experimental evidence is available (13).
Results
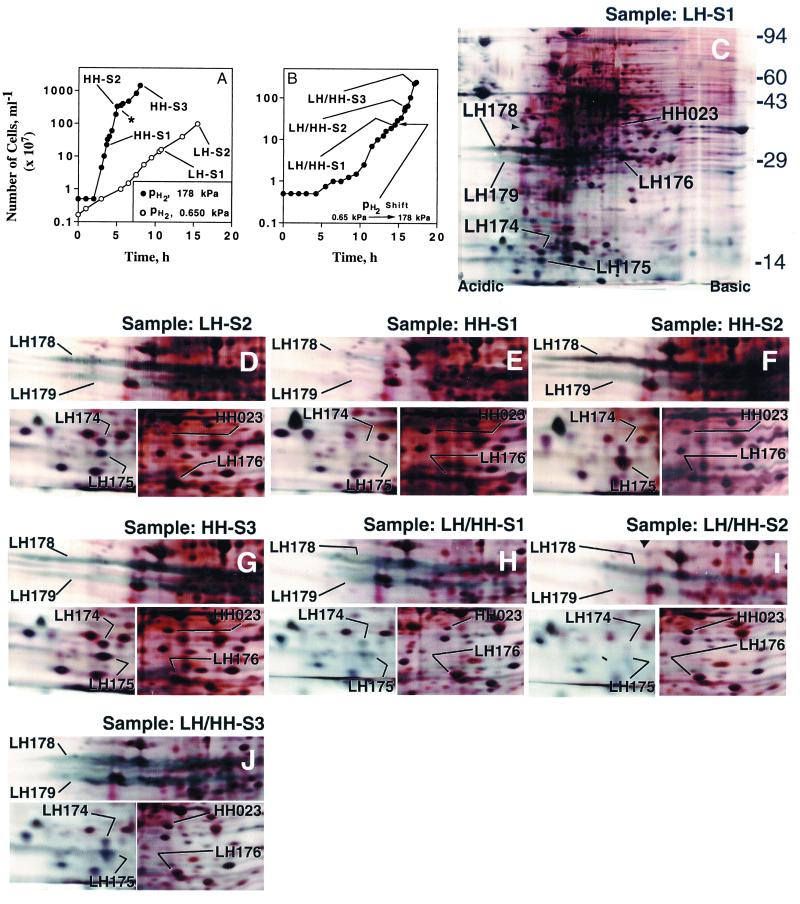
We studied autotrophic growth of M. jannaschii at two hydrogen partial pressures (pH2), 0.650 and 178 kPa, and henceforth these are referred to as low hydrogen or LH and high hydrogen or HH conditions, respectively. Fig. 1A shows the growth data for a LH and a HH culture and the collection points for the corresponding cell samples, and the 2-D gel electrophoresis patterns for these samples are in Fig. 1 C–G. We also conducted a pH2-shift experiment (Fig.1B) and analyzed the corresponding cell samples (Fig. 1 H–J). The following examples explain the nomenclature used in designating the cell samples: LH-S1, low hydrogen culture-Sample 1 (Fig. 1A); HH-S1, high hydrogen culture-Sample 1 (Fig. 1A); LH/HH-S1, low to high pH2-shift culture-Sample 1 (Fig. 1B).
Figure 1.
Growth of M. jannaschii in a reactor and 2-D gel electrophoretic analysis of cell extracts. (A) The growth data for a LH and a HH culture, and the collection points for the cell samples HH-S1, HH-S2, HH-S3, LH-S1, and LH-S2. The * points to the transition from a logarithmic to a linear growth phase (4). (B) A culture that was initiated at a pH2 of 650 Pa, but was switched later to 178 kPa; the culture samples were: LH/HH-S1, LH/HH-S2, and LH/HH-S3. (C–J) The 2-D gel patterns for the cell samples as indicated. The short arrow head in C points to the smaller isoform (33 kDa) of tropomyosin (pI, 5.2), an internal standard for an isoelectric focusing gel. The 2nd-dimension gel molecular-mass standards (kDa), whose locations are marked to the right of C, appear as indiscrete horizontal lines across a gel. For LH-S1 (C), the gel area including and below the 94-kDa standard is shown. For others, only three relevant sections (bearing the LH and HH spot areas) are shown. LH174, LH175, LH176, and HH023 appear as defined spots, whereas LH178 and LH179 are bluish streaks, indicative of their membranous origin.
The intensities of numerous 2-D gel spots were found to change with pH2. In this report, we focused our attention on six of those spots, and they belonged to two classes: (i) the low hydrogen or LH class, comprising LH174, LH175, LH176, LH178 and LH179; and (ii) the high hydrogen or HH class, represented by HH023.
2-D Gel Data for a Low pH2 or LH Culture.
As shown in Fig. 1A, two cell samples were collected from this culture: LH-S1 (cell density, 1.6 × 108 ml−1) and LH-S2 (cell density, 109 ml−1). Each of these samples exhibited high intensity LH spots (LH174, LH175, LH176, LH178, and LH179) and a very low intensity HH023 spot in the respective 2-D gel patterns (Fig. 1 C and D).
2-D Gel Data for a High pH2 or HH Culture.
The first sample, HH-S1 (cell density, 2.3 × 108 ml−1; Fig. 1A), exhibited a high intensity HH023 spot in the corresponding 2-D gel (Fig. 1E), but here the LH polypeptides were present at nondetectable or very low levels. However, the situation changed as the culture aged. The next sample, HH-S2, which was collected at a cell density of 3.4 × 109 ml−1 (Fig. 1A), exhibited LH174, LH175, LH178, and LH179 at high intensities (Fig. 1F) and HH023 with a reduced intensity (Fig. 1F); also, the intensity of LH176 increased, although not to a level that was seen for the LH culture samples LH-S1 and LH-S2 (Fig. 1 C and D). Thus, HH-S2 was similar to the LH culture samples. The gel pattern for HH-S3 (Fig. 1G), the third sample, was very similar to that for HH-S2 (Fig. 1F), although HH-S3 was collected from the linear growth phase (4) of the HH culture and at a cell density higher than that for HH-S2 (Fig. 1A).
The pH2-shift or LH/HH Experiment.
Fig. 1B shows the growth data for a culture that was initiated at a pH2 of 650 Pa, but was brought under a pH2 of 178 kPa at the early stage of growth (a very low cell density, 2.3 × 108 cells ml−1). The sample LH/HH-S1 was collected before the pH2 was increased (Fig. 1B), and the corresponding 2-D gel pattern (Fig. 1H) possessed high intensity LH spots (LH174–176, LH178, and LH179) and a low intensity HH023. Thus, it was similar to the LH culture samples LH-S1 or LH-S2 (Fig. 1 C and D). After the pH2 was increased 275-fold, the culture was allowed to progress for about a generation and then the sample LH/HH-S2 was collected (Fig. 1B). An analysis of this sample showed that the pH2-shift caused a transient decrease in the intensities of most LH polypeptides and an increase in the HH023 intensity (Fig. 1I); here, LH174 and LH175 were barely seen, LH178 intensity decreased, LH179 intensity decreased substantially, and LH176 intensity was comparable to that seen for LH/HH-S1 (Fig. 1H). But, as the culture progressed further to a cell density of 2.2 × 109 ml−1 (sample LH/HH-S3; Fig. 1B), the 2-D gel pattern began exhibiting very high amounts of LH174, LH175, LH178, and LH179, although the intensity for the LH176 and HH023 remained unaltered (Fig. 1J). Thus, the sample LH/HH-S3 was similar to HH-S2 and HH-S3, the late samples from the HH culture (comparing the data in Fig. 1J with that in Fig. 1 F and G).
The Identities for the LH and HH Polypeptide Spots in the 2-D Gel Patterns.
The results from mass spectrometry with the unseparated proteolytic digests of the polypeptide spots and database searches have been summarized in Table 1. From these, LH174, LH175, LH178, and LH179 were identified as follows, respectively: flagella-related protein D (FlaD), flagella-related protein E (FlaE), flagellin B2 (FlaB2), and flagellin B3 (FlaB3) (2, 13). Fig. 2 shows the relative locations of the genes for these polypeptides in the genome of M. jannaschii. The LH176 polypeptide was found to be the subunit of the F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD). The spot HH023 corresponded to the subunit of a putative H2-depedent methylenetetrahydromethanopterin dehydrogenase (HMDX; MJ0715); M. jannaschii possesses two more HMD homologs [HMD and HMDY or MJ0784 and MJ1338 (2)]. It was noted that only 14% of the sequence length of FlaD was represented by the peptide fragments of LH174 (Table 1). However, these fragments corresponded only to a C-terminal part (from and beyond residue 284) of FlaD (Table 1). Also, LH174 was ≈16 kDa in size, whereas the calculated mass value for FlaD was 39.95 kDa (Table 1 and Fig. 1 C–I). Therefore, the LH174 polypeptide was most likely a cleavage product of FlaD. If LH174 were to correspond to a ≈16-kDa C-terminal part of FlaD (viz., the residues 207–342), then the coverage value would be 27% and within an acceptable range (12). Also the theoretical pI of this fragment (a value of 5.14) would be similar to that for LH175 (Table 1), and in the isoelectric focusing or first dimension gels LH174 and LH175 comigrated. The coverage for the Fla polypeptides were in general a bit lower (Table 1), and this result could be indicative of protein modifications, such as glycosylation (14).
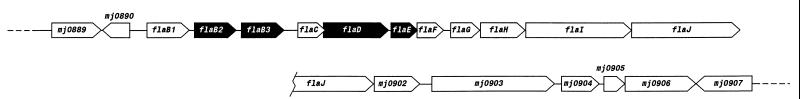
Figure 2.
Organization of putative flagella biosynthesis genes on the chromosome of M. jannaschii. The sequence of this region was determined by Bult et al. (2), and, wherever known, the gene names have been shown; these designations have been derived through comparisons to the fla genes of M. voltae (13). The FlaI protein might code for a type II secretion system protein. The filled arrow boxes represent those genes whose polypeptide products were located on the 2-D gels (Fig. 1).
Summary for 2-D Gel Data.
Here, we address the spots by their determined identities (Table 1). The polypeptides FlaB2, FlaB3, FlaD, and FlaE were either absent or of very low intensities in a 2-D gel pattern, only if the cell sample came from the logarithmic very low cell density (≈108 ml−1) phase of a HH culture. They were intense if the cells were from the mid- or late-logarithmic or the linear growth phase of a HH culture or from the logarithmic growth phase of a LH culture. The picture with MTD and HMDX was mixed. At the early stages of logarithmic growth, the higher pH2 condition favored an elevated level of HMDX and a reduced level of MTD, and the lower pH2 condition gave the exact opposite effect. But, when a HH culture aged, both HMDX and MTD were found at high intensities. All these observations were reconfirmed by repeat 2-D gel analyses for the above described samples, as well as by analyzing samples from duplicate cultures (data not shown).
Electron Microscopy.
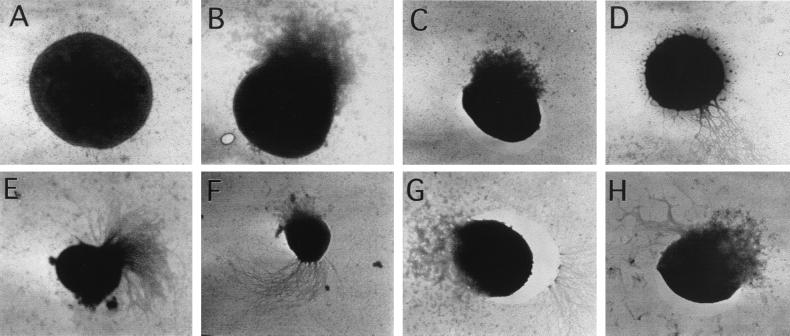
We examined the cells from the logarithmic stage of a HH culture (Table 2) for the extent of flagellation by use of transmission electron microscopy. Fig. 3 A–H shows the electron micrographs for various types of stained cells that were observed under the microscope. Often a cell was seen burst open at one side (Fig. 3 B and C), and, for flagellated cells, this damage occurred almost exclusively at the side opposite to that where the flagella were seen (Fig. 3F); M. jannaschii possesses pseudopolar flagella [“two waved-bundles … inserted close to the same cell pole” (1)]. Hence, such damage to a cell did not obscure the fact that a cell was flagellated or nonflagellated. In rare instances, damage occurred at the flagella side (Fig. 3H), but the flagella and cell debris were distinguishable. Fig. 3D shows a very rare weakly flagellated cell from the sample EM1. The data in Table 2 show that, at the very early stage of a HH culture (samples EM-1 and EM-2), almost each and every cell was devoid of flagella. However, as the culture progressed, yet remaining at the logarithmic stage, most cells were seen to have flagella (Table 2). In the last sample (EM-5), about 95% of cells were flagellated.
Table 2.
The changes* in the number of flagellated cells in a logarithmic phase reactor culture of M. jannaschii operated at a pH2 of 178 kPa
| Sample number | Number of cells per ml culture | Number of flagellated cells†‡§ | Number of nonflagellated cells†§ | % of the population flagellated |
|---|---|---|---|---|
| EM-1 | 8 × 106 | 3 | 102 | 2.86 |
| EM-2 | 1.7 × 108 | 0 | 126 | 0 |
| EM-3 | 5 × 108 | 61 | 38 | 61.6 |
| EM-4 | 1.6 × 109 | 147 | 36 | 80.3 |
| EM-5 | ∼4 × 109 | 364 | 18 | 95.3 |
Transmission electron microscopic observation of negatively stained cells.
Cells were counted in randomly selected fields; an arbitrarily number of cells was counted.
No distinction was made based on the extent of flagellation.
See Fig. 3 for cell types.
Figure 3.
Electron micrographs of negatively stained M. jannaschii cells. The cells were from the following samples that were collected from the logarithmic phase of a reactor culture raised at a pH2 of 178 kPa (see Table 2). (A–D) Sample EM-1; (E–H) sample EM-4. Magnifications (×103): (A) 10.5; (B) 9.4; (C) 5.4; (D) 5.5; (E) 5.6; (F) 5.5; (G) 8.3; and (H) 6.8. To generate the data in Table 2, cell types A, B, and C were scored as nonflagellated; D, E, F, G, and H were scored as flagellated.
Discussion
This report presents the first example for experimental functional genomics and proteomics for M. jannaschii. Here, the effect of an environmental factor on the organism's behavior was linked to a region of the genome, and the experiments were conducted with no prior knowledge of this linkage.
The observed well-concerted changes in the levels of four Fla polypetides, along with corroboration by the data from microscopic observation, clearly indicated the existence of a control on the flagella synthesis by pH2 in M. jannaschii. To our knowledge, there have been no other reports about the regulation of expression of flagella by hydrogen in any domain of life and for a control of any kind on flagella synthesis in the archaea. If these archaeal appendages are considered as type IV pili (Tfp), as suggested (13), then it would be true that our results helped to identify for the first time a chemically defined nutritional signal for the expression of a Tfp homolog. This observed regulation was not a cell density-dependent phenomenon, because cells from low density LH cultures (Figs. 1 C, D, and H), as well as from high density HH cultures (Fig. 1 F, G, and J), exhibited high intensity Fla polypeptides. An operon-like arrangement for the M. jannaschii fla genes (Fig. 2) fits well the fact that the levels of four Fla polypeptides were coregulated. Indeed, a polycistronic fla message covering the region from flaB1 to flaG (or flaI) has been found in Methanococcus voltae (13, 15), a close relative of M. jannaschii. In M. voltae, FlaB2 is the major flagellin, and FlaB3 is considered a minor flagellin (14). The M. jannaschii FlaD and FlaE appeared as smears in the 2-D gels (Fig. 1 C, D, and F–H and 1J). Thus, they might belong to a membrane component of the flagella or Tfp apparatus. These observations explain why FlaB3, FlaD and FlaE have not been detected as components of purified methanococcal flagella filaments (14).
Noting its chemoautotrophic nature and the absence of decipherable chemotaxis genes in its genome, Faguy and Jarrell (16) speculated that M. jannaschii is probably not chemotactic, but only thermotactic. Our data clearly contradict that hypothesis and show that this archaeon does make a connection between a chemical signal (pH2) and the expression of a motility apparatus. This discovery sets the stage for a search for the chemotaxis systems in M. jannaschii. The hydrogen concentration in vent emissions and the surroundings is highly variable and lies in the 0.03 μM-1.7 mM range (pH2, 4 Pa to 233 kPa) (5). Thus, an ability to control flagella biosynthesis and hence locomotion (both being rather energy-intensive processes) by pH2 would poise this strict hydrogenotroph, which has a poor energy-yielding pathway (17), to make best use of the environmental resources.
It was only in the very early logarithmic and low cell density phase of a HH culture where the cells had low or unobservable levels of flagella and Fla polypeptides. The sample HH-S2 was collected at a culture cell density of only 3.4 × 109 ml−1, and yet it had high levels of Fla polypeptides. Thus, M. jannaschii was capable of sensing subtle changes in dissolved hydrogen concentration. Hence, an opportunity exists for discovering an exceptionally sensitive, efficient, and fast-acting signal-sensing and signal-transmission system in this archaeon.
The nature of the hydrogen regulation of MTD and HMDX proteins in M. jannaschii was very similar to that seen for the mtd, hmdI, and hmdIII transcripts in Methanobacterium thermoautotrophicum strain ΔH (18, 19). Thus, the observed regulation of the Fla polypeptide's synthesis paralleled a previously characterized H2-level-depenedent behavior. Hence, MTD and HMDX acted as the internal controls. However, it remains unclear why the control on the expression of mtd and hmds by pH2 is apparently lost as a culture ages (refs. 18 and 19 and this work); here, both mtd and hmd genes are expressed at high levels.
It is worth noting that the pH2values in common freshwater methanogenic habitats fall in the 1–2000 Pa range (20), and the need to consider pH2 values of 1–10 Pa, when evaluating H2-dependent methanogenesis data, has been stressed (17). The pure-culture cultivation of a methanogen at an ultra-low or near ecologically relevant pH2 value has not been reported. Our LH cultures approached these conditions.
Acknowledgments
We thank Melody Kyper, Jon Johansen, and Nancy Kendrick for consultation on 2-D gel elctrophoresis; Mary Ann Gawinowicz for help in deciphering the identities of the polypeptide spots on 2-D gels; Eric J. Chaney and Betty Ujhelyi for assisting in electron microscopy; and Endang Purwantini for discussions. All reactor culture experiments were conducted in the Department of Microbiology Fermentor Facility, University of Illinois at Urbana-Champaign. This work was supported by Department of Energy Grant DE-FG02-87ER13651 and National Institutes of Health Grant GM51334.
Abbreviations
- LH
low hydrogen
- HH
high hydrogen
- MTD
F420-dependent methylenetetrahydromethanopterin dehydrogenase
- HMDX
H2-dependent methylenetetrahydromethanopterin dehydrogenase homolog
References
- 1.Jones W J, Leigh J A, Mayer F, Woese C R, Wolfe R S. Arch Microbiol. 1983;136:254–261. [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Tumbula D L, Whitman W B. Mol Microbiol. 1999;33:1–7. doi: 10.1046/j.1365-2958.1999.01463.x. [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay B, Johnson E F, Wolfe R S. Appl Environ Microbiol. 1999;65:5059–5065. doi: 10.1128/aem.65.11.5059-5065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jannasch H W, Mottl M J. Science. 1985;229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- 6.Glauert A M. Fixation, Dehydration and Embedding of Biological Specimens. New York: North–Holland; 1975. [Google Scholar]
- 7.Basgall E J, Scherba G, Gelberg H B. Proceedings: Electron Microscopy Society of America, Forty-sixth Annual Meeting. San Francisco: San Francisco Press; 1988. pp. 366–367. [Google Scholar]
- 8.O'Farrell P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connell K L, Stults J T. Electrophoresis. 1997;18:349–359. doi: 10.1002/elps.1150180309. [DOI] [PubMed] [Google Scholar]
- 10.Rosenfeld J, Capdevielle J, Guillemot J C, Ferrara P. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 11.Sheer D G. In: Techniques in Protein Chemistry. Crabb J W, editor. V. San Diego: Academic; 1994. pp. 243–248. [Google Scholar]
- 12.Mann M, Hojrup P, Roepstorff P. Biol Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- 13.Bayley D P, Jarrell K F. J Mol Evol. 1998;46:370–373. [PubMed] [Google Scholar]
- 14.Jarrell K F, Bayley D P, Kostyukova A S. J Bacteriol. 1996;178:5057–5064. doi: 10.1128/jb.178.17.5057-5064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalmokoff M L, Jarrell K F. J Bacteriol. 1991;173:7113–7125. doi: 10.1128/jb.173.22.7113-7125.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faguy D M, Jarrell K F. Microbiology. 1999;145:279–281. doi: 10.1099/13500872-145-2-279. [DOI] [PubMed] [Google Scholar]
- 17.Thauer R K. Biochim Biophys Acta. 1990;1018:256–259. [Google Scholar]
- 18.Morgan R M, Pihl T D, Nolling J, Reeve J N. J Bacteriol. 1997;179:889–898. doi: 10.1128/jb.179.3.889-898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeve J N, Nolling J, Morgan R M, Smith D R. J Bacteriol. 1997;179:5975–5986. doi: 10.1128/jb.179.19.5975-5986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinder S H. In: Methanogenesis: Ecology, Physiology, Biochemistry, and Genetics. Ferry J G, editor. New York: Chapman & Hall; 1993. pp. 128–206. [Google Scholar]