Abstract
A microtubule-associated protein, γ-aminobutyric acid type A (GABAA) receptor-associated protein (GABARAP), was previously identified as binding to the intracellular domain of GABAA receptors by using the yeast two-hybrid screen. In the present work, immunofluorescent staining and green fluorescent protein-tagged receptor subunits showed that GABARAP is associated with and promotes the clustering of GABAA receptors in QT-6 quail fibroblasts. The tubulin-binding motif of GABARAP and the γ2 subunit of the receptor are required. Disruption of microtubules prevents the clustering in a time-dependent manner. When green fluorescent protein-tagged α1 or γ2 subunit coexpressed with β2, γ2L, and GABARAP was used, recordings from visually identified cells revealed that clustered GABAA receptor had an EC50 of about 20 μM, vs. 5.7 μM for the diffuse receptor. Clustered receptors deactivated faster and desensitized slower than the diffuse receptors, because of decrease in the apparent affinity of GABA binding. Different properties for clustered receptors relative to unclustered receptors in heterologous cells suggest that homologous differences between extrasynaptic and synaptic clustered receptors in neurons may be due to the organization of the postsynaptic machinery.
Targeting and clustering of γ-aminobutyric acid type A (GABAA) receptors to specific membrane areas are crucial for their normal function. For fast synaptic transmission, GABAA receptors must be clustered underneath GABAergic termini (1). This clustering presumably reflects anchoring to the cytoskeleton, and disruption of the cytoskeleton affects some pharmacological properties of GABAA receptors (2, 3). Decreased GABAA receptor clustering results in dysfunction of GABAA receptors at the cellular level and anxiety disorders at the animal level (4). Gephyrin was reported to colocalize with GABAA receptor at synaptic sites in retina and spinal cord (5, 6). In GABAA receptor γ2 subunit knockout mice, clusters of GABAA receptors were decreased greatly, and most of the gephyrin staining was also gone (7). But a direct interaction between gephyrin and GABAA receptors has not been proved so far (8, 9). The 43-kDa protein rapsyn, associated with nicotinic acetylcholine receptors at the neuromuscular junction (10), was found to cluster with GABAA receptors when coexpressed in QT-6 cells (11), but its very low level of expression in the brain excludes rapsyn as a major component for clustering GABAA receptors. Recently our group cloned a microtubule-binding protein, GABAA receptor-associated protein (GABARAP), which is a putative linker protein between cytoskeleton and GABAA receptors (12, 13).
Here, we report a functional assay for GABARAP. Immunofluorescent staining and green fluorescent protein (GFP)-tagged receptor subunits show that GABARAP promotes GABAA receptor clustering. Patch clamp studies reveal that clustering changes the channel kinetics of the GABAA receptors.
Materials and Methods
Cell Culture, Transfection, and Immunofluorescent Staining.
Japanese quail QT-6 fibroblasts were grown and transfected with the method described by Yang et al. (11). The plasmids (pCDNA3) encoding α1, β2, and γ2L GABAA receptor subunits were made in the laboratory of R.W.O. (12). Full-length GABARAP cDNA including 5′ and 3′ untranslated regions was amplified by PCR and cloned into pCR II (Invitrogen). Then the GABARAP insert was removed from pCR II by EcoRV and HindIII digestion, treated with the Klenow fragment of DNA polymerase, and cloned into the EcoRV site of pCDNA3 (Invitrogen). Two resultant plasmids were generated. One, pCDNA3-GABARAP, will transcribe GABARAP mRNA under the control of the cytomegalovirus (CMV) promoter. The other, pCDNA3-GABARAP/antisense, will transcribe RNA complementary to GABARAP mRNA. Similarly, pCDNA3-GABARAP/36–117 was generated to produce the truncated form of GABARAP. Plasmid DNA for GABAA α1 subunit with GFP tags was made in the laboratory of S.V.‡‡ γ2L-GFP in pRK5 vector was a gift from Stephen J. Moss (University College, London).
Seventy-two hours after transfection the cells on glass coverslips were fixed for 30 min and stained with anti-β-chain antibody bd17 by using an immunofluorescent staining method (11). The fluorescence microscopy was performed on a Nikon Microphot-FXA microscope with a 60× oil-immersion objective. For GABAA receptor and GABARAP double-label experiments, QT-6 cells were permeabilized with 1% Triton X-100 in PBS after bd17 staining, then incubated with GABARAP antibody for 1.5 h at room temperature. After washing with PBS, coverslips were incubated for 1.5 h with fluorescein-conjugated horse anti-mouse IgG (1:100; Vector) and Texas red-conjugated goat anti-rabbit IgG (1:100; Vector). After washing, the coverslips were mounted for confocal fluorescence microscopy. To verify the involvement of microtubules in clustering of GABAA receptors, overnight treatment with the microtubule disruption agent nocodazole (1 μg/ml) was initiated at various times after transfection with α1, β2, and γ2L-GFP GABAA receptor subunits and GABARAP. The GFP-tagged receptors were analyzed with fluorescence microscopy.
Rabbit polyclonal antibody was generated against glutathione S-transferase (GST)-GABARAP. The GABARAP antibody was purified from the crude serum by antigen affinity column using Affi-Gel 10 (Bio-Rad) coupled with GST-GABARAP (12).
Electrophysiology.
QT-6 cells transfected with α1-GFP(pEGFP-α1), β2(pCDNA3), γ2L(pCDNA3), and GABARAP(pCDNA3) were treated with collagenase I (Sigma) at 1.5 mg/ml in 1 mM CaCl2 external solution for 30 min at room temperature. The cells were dissociated with fire-polished glass Pasteur pipettes and transferred to the recording chamber in a recording external solution containing (in mM): 140 NaCl, 4.7 KCl, 1.2 MgCl2, 2.5 CaCl2, 11 glucose, and 10 Hepes (pH 7.4). Then the cells were examined under a fluorescence microscope (Nikon Eclipse TE200). The GABAA receptors were visualized by GFP fluorescence at 510 nm. The GFP-tagged GABAA receptors were shown to be expressed and to produce normal channel properties and pharmacology by Vicini et al.†† and as also shown by other groups (14). Whole-cell patch clamp recordings were made from visually identified cells with clustered or diffuse GABAA receptors (Fig. 3). Patch pipettes were made from borosilicate glass capillaries (Sutter Instruments, Novato, CA) and filled with an internal solution containing (in mM): 125 CsMeSO3, 15 CsCl, 10 glucose, 10 Hepes, 2 EGTA, and 5 MgATP (pH 7.2).
Figure 3.
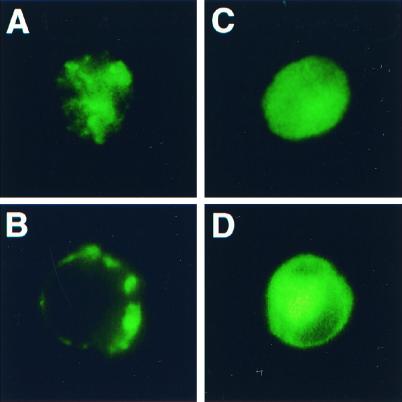
GFP-tagged GABAA receptor subunits visualize the receptors in live QT-6 cells. The cells shown in A and B were transfected with plasmids encoding α1-GFP, β2, γ2L, and GABARAP. After 72 h of expression, the cells were dissociated and replated into a recording chamber and the picture was taken with a Nikon inverted fluorescence microscope. A shows the clustered receptors on the top of the cell. B shows broken perimeter indicative of clusters. C and D were taken from cells expressing α1-GFP, β2, γ2L and without exogenous GABARAP. These pictures show diffuse receptors.
To study kinetics of GABA responses we choose to use whole-cell recordings to prevent alteration of essential cytoskeletal components. In this experimental condition it is impossible to obtain a sufficiently rapid exchange rate to assess rapid components of desensitization (15). Our estimates of desensitization and deactivation reflect this limitation. In this study, GABA was applied to the recorded cell by means of the “concentration clamping” fast perfusion system (16). The junction potential measurements indicate that the solution exchange was accomplished in 6 ms. Because the fast components of desensitization become significant only at high concentrations (>100 μM) (17), we can avoid fast desensitization by using a low concentration of GABA (<100 μM) to measure the rate constants of desensitization and deactivation. In the case of EC50 measurement, even at a concentration of 10 times EC50 (50–200 μM), the fast component of desensitization (τ = 15–20 ms) had little effect on peak currents (<10%) in our system, so it has little influence on EC50 value. The currents were recorded with a patch clamp amplifier (Axopatch 200B) and digitized with DigiData 1200 at 1 ms per point. The data were sampled and processed with PCLAMP-7 (Axon Instruments, Foster City, CA).
Statistics.
Whole-cell currents were analyzed off-line with clampfit (Axon Instruments) and prism (GraphPad Software, San Diego). The dose–response data were fit with a four-parameter logistic equation: I = Imax/{1 + 10(log EC50 − log[GABA])H)}, where Imax is the maximum current induced by saturating dose of GABA, the Hill coefficient H is 1.0, and [GABA] is the concentration of GABA. The t test was performed to measure statistical significance.
Results
GABARAP Promotes GABAA Receptor Clustering.
GABARAP and GABAA receptor subunits α1, β2, and γ2L were coexpressed in the quail fibroblast cell line QT-6. The expressed GABARAP was detected by Western blotting of cell homogenates (particulate fraction). Interestingly, QT-6 cells had only a trace background expression of endogenous GABARAP; compared with other cell lines such as Sf-9 cells and with rat brains, the expression level was very low (data not shown). QT-6 cells in culture usually have two types of morphologically different cells: large flat cells with long processes and spherical cells with few processes. The GABAA receptors were visualized by immunofluorescent staining and confocal fluorescence microscopy. To show only the receptors on the membrane, the cells were not permeabilized before primary antibody staining with bd17, which recognizes the GABAA receptor β2 subunit. Fig. 1A shows a large flat cell expressing GABARAP plus α1β2γ2L GABAA receptors. The immunohistochemically detected GABAA receptors were clustered on the cell membrane. The spherical cell in Fig. 1B expressed GABARAP, and GABAA receptors were also clustered on the membrane (note the “broken” periphery). The cell shown in Fig. 1C was transfected with antisense GABARAP plus α1β2γ2L GABAA receptors. This cell showed a diffuse receptor staining without clusters. Fig. 1 D, E, and F are confocal images of cell surface GABAA receptors (green fluorescence: antibody stained before permeation) and GABARAP (red fluorescence: stained following permeation). Fig. 1F is the merge of 1D and 1E; yellow indicates colocalization of GABAA receptors and GABARAP.
Figure 1.
GABARAP cluster GABAA receptors. This figure shows immunofluorescence of GABAA receptor expressed in QT-6 cells with or without GABARAP. All of the cells shown here expressed α1β2γ2L GABAA receptor and stained with antibody bd17. (A) Large flat cell expressing GABARAP gives clustered GABA receptors. (B) Spherical cell with GABARAP gives clusters; note “broken” periphery. (C) Large flat cell with antisense GABARAP shows diffuse GABAA receptors. (D and E) Confocal images of antibody-labeled GABAA receptors (D, green) and GABARAP (E, red) at the cell surface. (F) A merge of D and E, showing colocalization in yellow.
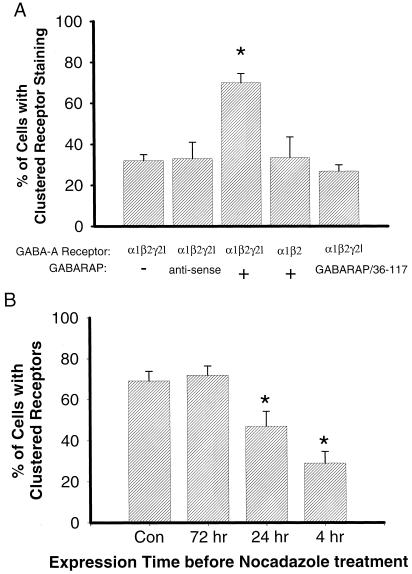
To quantify the effect of GABARAP, we randomly grouped every 100–120 bd17-positive cells into a group, then counted the percentage of cells with clustered staining in each group [(number of cells showing clustered stain/total of the group) × 100%]. We found that only about 30% of cells showed clustered staining in control groups (α1β2γ2L, 31.9% ± 3%, mean ± SD, n = 6) or control (α1β2γ2L) plus antisense GABARAP (32.9% ± 8%, n = 8), but in the groups expressing α1β2γ2L + GABARAP, 70% of the cells showed clustered staining (70% ± 4.5%, n = 10) (Fig. 2A). The difference between control and GABARAP groups is statistically significant (P < 0.001, t test). On the basis of these data, we concluded that GABARAP promotes GABAA receptor clustering.
Figure 2.
The γ2 subunit, tubulin-binding motif of GABARAP, and microtubules are involved in clustering of GABAA receptors. (A) GABARAP promotes GABAA receptor clustering when coexpressed in QT-6 cells. The bar graph summarizes the percentage of cells that had GABAA receptor clusters. The expressed subunits and GABARAP are shown below. The bd17-positive QT-6 cells were randomly divided into about 100 cells per group, then the percentage of cells with clustered staining in each group was counted [(number of cells showing clustered stain/total of cells in the group) × 100%]. Each bar represents the mean and SD of 6–10 groups of the cells. *, P < 0.001 (t test). (B) Microtubule disruption prevents clustering of GABAA receptors in a time-dependent manner. QT-6 cells were transfected with plasmids encoding α1, β2, γ2L-GFP GABAA receptors and GABARAP. The microtubule disruption agent nocodazole (1 μg/ml) was added into culture medium at 4 h, 24 h, and 72 h after transfection. GFP-tagged receptors were examined with clustering assay described above. Each bar represents the mean and SD of 4–6 groups of the cells. *, P < 0.001 (t test).
The γ2 Subunit and the N-Terminal Tubulin-Binding Motif of GABARAP Are Required for Clustering of GABAA Receptors.
We have shown previously (12) that GABARAP specifically binds to the γ2 subunit, and its N-terminal 36 amino acid sequence is not required for receptor interaction, but contains a tubulin-binding motif. Using the QT-6 cell line, we can express various receptor subunits, subunit combinations, or mutated subunits. We can also coexpress truncated or mutant GABARAP to study functional domains. In the cells that expressed α1 and β2 but not γ2L subunits with GABARAP, only 33% of the cells showed clustered staining, which is not different from control groups (α1β2γ2L without GABARAP, Fig. 2A). We also tested a truncated version of GABARAP, GABARAP(36–117), which lacks the putative tubulin-binding motif. When GABARAP(36–117) was coexpressed with α1β2γ2L subunits, most of the cells' GABAA receptors showed a diffuse pattern of distribution. GABARAP(36–117) had no effect on the clustering of GABAA receptors (Fig. 2A). These clustering assay results indicate that the γ2 subunit and tubulin-binding motif are required for GABARAP action. GABARAP may anchor and cluster GABAA receptors through the γ2 subunit and the tubulin-binding motif, which binds to the cell skeleton.
Disruption of Microtubules Prevents the Clustering in a Time-Dependent Manner.
GFP-tagged α1 or γ2 subunits were expressed in α, β, γ combinations to visualize GABAA receptors (Fig. 3). This figure shows that in the presence of GABARAP, GABAA receptors show clusters at the cell surface (Fig. 3A) with a broken perimeter (3B) in the majority of the cells, whereas the receptors are diffusely expressed at the surface (3C) with unbroken perimeter (3D) in the absence of GABARAP.
To verify the involvement of microtubules in clustering of GABAA receptors, overnight treatment with the microtubule disruption agent nocodazole (1 μg/ml) was initiated at various times after transfection, and GFP-tagged receptors were examined at 84 h by using the clustering assay mentioned above. We found about equal percentage of the labeled cells showing clustered receptors in both control (69% ± 4.7%, mean ± SD) and 72-h groups (72% ± 4.5%). But early treatment with nocodazole can prevent receptor clustering at the cell membrane. In the 24-h group, 47% ± 7.2% of cells showed clusters, and in the 4-h group, only 29% ± 5.7% showed clusters (Fig. 2B). Immunofluorescent staining with anti-β-tubulin antibody in QT-6 cells revealed that microtubules were diffusely distributed throughout the cells, with no punctate staining on the membrane (data not shown). These results indicate that microtubules may be involved in GABAA receptor trafficking instead of physically holding the receptors together.
Clustering of GABAA Receptors Decreases Apparent Affinity for GABA.
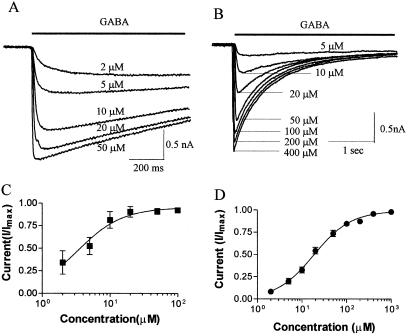
Coexpressing α1-GFP with β2, γ2L, and GABARAP in QT-6 cells, we were able to see the distribution pattern of GABAA receptors in live cells (Fig. 3) and to make recordings from visually identified cells. The cells we chose exhibited the vast majority if not all of the GABAA receptors in either the diffuse or clustered state. Both sorts of cells responded to GABA and benzodiazepines, consistent with the αβγ subunit composition. Using a fast perfusion system and whole-cell patch clamp technique, we measured the apparent affinity (EC50) of GABA for diffuse and clustered receptors (Fig. 4). The EC50 value was 5.74 ± 1.4 μM (n = 7) for diffuse receptors, but it increased to 20.27 ± 3.8 μM (n = 7) for clustered receptors. The difference in EC50 is statistically significant (P < 0.005, t test).
Figure 4.
Clustering of GABAA receptors decreases apparent affinity for GABA. (A) Whole-cell patch clamping recordings of GABA-induced currents from diffuse GABAA receptors. The line above the traces indicates the time period of GABA application. The holding potential was −60 mV. The GABA responses saturated at 50–100 μM. (B) Whole-cell currents from a cell with clustered GABAA receptors. The holding potential was −60 mV. The peak currents saturated at 400-1000 μM GABA. (C) Dose–response curves for diffuse GABAA receptor. The data were fit with a four-parameter logistic equation: I = Imax/{1 + 10(log EC50 − log[GABA])H)}, where Imax is the maximum current induced by saturating dose of GABA, the Hill coefficient H is 1.0, and [GABA] is the concentration of GABA. From seven cells, the average EC50 is 5.7 ± 1.4 μM (mean ± SE). (D) Dose–response relationship of clustered GABAA receptors. The data were fit with the same equation as in C. From seven cells, the average EC50 is 20.3 ± 3.8 μM (mean ± SE). The large difference in apparent affinity between clustered and diffuse receptors is statistically significant (P < 0.001, t test).
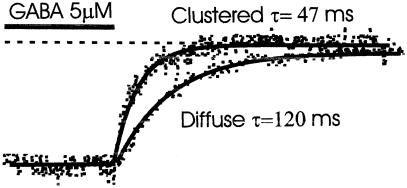
To further investigate the mechanism of the affinity change, we studied the deactivation rate of GABAA receptor channels in clustered or diffuse state (Fig. 5). The deactivation time constant τ was measured after 4-s application of 5–10 μM GABA. The deactivation time constant was much longer in the diffuse state than in clusters (117.6 ms vs. 51 ms). In other words, clustered receptors deactivate much faster. That may explain why the GABAA receptors clustered underneath presynaptic terminals generate inhibitory postsynaptic potentials shorter than those produced by extrasynaptic diffuse receptors (18–20). We also tested the I/V relationship of diffuse and clustered GABAA receptors. They had similar I/V curves and almost identical reversal potentials (−38.0 ± 1.7 mV vs. −39.4 ± 2.0 mV, mean ± SD, n = 3, n = 5 respectively).
Figure 5.
Clustering increases deactivation rate. The cell was held at −60 mV, and the dashed line indicates zero current level. GABA (5 μM) induced an inward current. When GABA was rapidly removed, the current gradually decreased to zero. The deactivation phase of currents was best fit with a single exponential. Diffuse receptors had an average time constant τ = 117 ± 10.5 ms (mean ± SE, n = 6); clustered receptors' deactivation rate was faster, with time constant shorter, τ = 51 ± 4 ms (mean ± SE, n = 3). The difference is statistically significant (P < 0.05, t test).
Clustering of GABAA Receptors Decreases Desensitization.
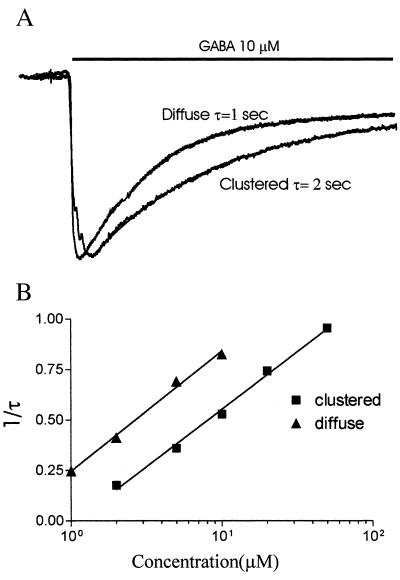
In the cells expressing α1-GFP, β2, and γ2L GABAA receptors, the desensitization time courses for low GABA concentrations can be best fit by a single exponential (Fig. 6A). The time constant (τ) decreased when GABA concentration was increased. For the same concentration of GABA, clustered receptors desensitized slower than diffuse receptors (Fig. 6A). For example, in the presence of 10 μM GABA, the diffuse receptors desensitized with a τ value of 1 s. But in clustered receptors, τ was about 2 s (Fig. 6A). Plotting τ against GABA concentration, we found that in the low concentration range (1–10 μM for diffuse receptor, 2–100 μM for clustered receptor), τ had obvious concentration dependence. In the high concentration range, because fast desensitization kicked in, it was beyond the capability of our system to measure τ reliably (details in Materials and Methods). Plotting 1/τ vs. log[GABA] in the low concentration range, we found 1/τ had a linear relationship with log[GABA]. The data can be fit with a linear equation: 1/τ = α log[GABA] + C. Interestingly, diffuse receptors and clustered receptors have almost identical α but quite different C values. Compared with diffuse receptors, clustered receptors just parallel-shift the line to the right (Fig. 6B). This result suggests that clustering may change not the concentration-dependent rate constant of desensitization but the apparent affinity of GABA binding. That is consistent also with the dose–response curves (Fig. 4).
Figure 6.
Clustering changes desensitization. (A) Two traces of 10 μM GABA-induced currents were normalized. The line above the traces indicates the time period of GABA application. The holding potential was −60 mV. The decay of the currents was fitted with a single exponential. Diffuse receptors desensitized at a time constant τ = 1 s; clustered receptors had a τ = 2 s. (B) The concentration-dependent rate constant of desensitization. The data points represent the average value from 5 cells with diffuse receptors and 7 cells with clustered receptors. The data were fitted with an equation: 1/τ = α log[GABA] + C, where τ is the time constant for desensitization, [GABA] is the concentration of GABA, α is the rate constant of desensitization, C is the desensitization rate (1/τ) at [GABA] = 1 μM. Both diffuse and clustered receptors had almost same α value (0.57 vs. 0.59 M−1⋅s−1) but different C (0.25 vs. 0 μM). This result means that clustering may change not the concentration-dependent rate constant of desensitization, but the apparent affinity of GABA binding.
Discussion
GABARAP Clusters GABAA Receptors.
Immunofluorescent staining shows that GABARAP promotes GABAA receptor clustering. Recombinant α1, β2, and γ2L receptors coexpressed in QT-6 cells with GABARAP, but not with antisense GABARAP, show clustering to a significantly greater extent than in the absence of exogenous GABARAP. Receptors expressed with these three subunits contain γ2 subunits as indicated by the sensitivity to benzodiazepines and by observation of similar clustering behavior for receptor tagged with GFP on either α1 or γ2 subunit. These cells have a low level of endogenous immunoreactivity for GABARAP, which may account for the low degree of clustering seen with GABAA receptors expressed without GABARAP. The receptor γ2 subunit is required and the tubulin-binding domain in GABARAP is also essential for the effect: GABAA receptor α1 + β2 subunits express channels but are not clustered by coexpression with GABARAP. Truncated GABARAP lacking the N-terminal 35 residues does not cluster receptors. Our data indicate that GABARAP may cluster GABAA receptor by linking the intracellular domain of γ2 subunits. This linking can contribute to the intracellular scaffolding required for formation of postsynaptic receptor clusters. Clustering of GABAA receptors through γ2 subunits linked by GABARAP to the cytoskeleton is consistent with observations on γ2 knockout mice, in which most of the GABAA receptor clusters disappear in γ2 −/− homozygous null mutants, and the clustering is greatly decreased in γ2 +/− heterozygotes (4, 7). Disruption of microtubules with nocodazole at early times in GABAR-GABARAP coexpression prevented the formation of the clusters, indicating a requirement for cytoskeleton in clustering. At late times when clusters were already formed, disruption of microtubules did not lead to loss of clusters. This result is consistent with recent observations by Allison et al. (21) on real neurons.
Clustering Produces Unique Kinetics: Clusters Have Properties Consistent with Those of Native Synaptic Transmission.
The size and duration of excitatory and inhibitory postsynaptic potentials are largely determined by transmitter release and postsynaptic channel kinetics (15, 22–24). Modulation of kinetics is an important mechanism to modulate synaptic transmission (25–28). Many drug actions are based on this concept (29–34). There are several ways to modulate channel kinetics, such as changing subunit composition (17, 35), phosphorylating channel proteins (28, 36–39), binding of modulator to specific modulatory sites (e.g., the glycine site for N-methyl-d-aspartate receptor and neurosteroid binding site for GABAA receptor), etc.
In this study, we report that clustering of GABAA receptors can affect their kinetics. The apparent affinity, deactivation rate, and concentration dependence of desensitization all changed after clustering. This report presents evidence that one important factor in determining kinetics of ligand-gated ion channel properties is clustering of the receptors in the postsynaptic membranes, mimicked here by clustering in heterologous cells coexpressing GABARAP and GABAA receptors, including the γ2 subunit. Anatomical and physiological evidence (1, 19, 20, 40, 41) supports the presence of synaptic receptors positioned in clusters apposed to presynaptic terminals, but also extrasynaptic receptors with different properties. Extrasynaptic receptors are believed to respond to synaptic spillover of neurotransmitter (19, 42), and they must be capable of responding to lower concentrations. Currents ascribed to synaptic receptors were found to have more rapid deactivation than did those of extrasynaptic receptors (20).
The apparent affinity for GABA is lower for clustered receptors. This finding suggests that protein–protein interactions of subunits by means of intracellular domains can affect extracellular ligand binding site conformation, or coupling between ligand binding site and channel activation. This lower apparent affinity for GABA of clustered GABAA receptors apparently also translates into faster deactivation and slower desensitization at any given low GABA concentration. The apparent lower affinity and faster dissociation rate would be adequate for faster channel closing upon agonist deoccupancy. Deactivation rates measured are consistent with literature values for GABAA (36, 43, 44) and glutamate or nicotinic receptors (21, 45, 46). These deactivation rates are slow (50–100 ms) compared with the time required to remove GABA from the recording chamber (<10 ms), so the differences measured are highly significant. Our measurement of deactivation rate was made at a concentration of agonist that produced minimum desensitization, but we cannot rule out some contribution of desensitization. These studies do not measure very early events after application or removal of agonist because of technical difficulties (see Materials and Methods). The deactivation and desensitization rates measured thus do not reflect very rapid events (15), but show significant differences between clustered and diffuse receptors for the slow events. The difference in deactivation and desensitization may be accounted for by the change in agonist binding affinity, although a difference in channel properties is possible. Further experiments will be required to test this idea.
The cellular distribution of receptors into aggregates appears to involve both self-interaction and interactions with other gene products. At synapses especially, development of the postsynaptic membrane involves a complex scaffolding of many proteins, regulated by cell–cell signaling (47). Some aggregates of receptors are observed in the absence of presynaptic connection, but the organization of synaptic clusters is much more elaborate and specific, and thus important for synaptogenesis and plasticity.
Acknowledgments
National Institutes of Health Grants NS28772 and NS35985 to R.W.O. and National Institutes of Health Training Grant HD06576 to L.C. support this work.
Abbreviations
- GABA
γ-aminobutyric acid
- GABAA receptor
type A GABA receptor
- GABARAP
GABAA receptor-associated protein
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 11135.
Vicini, S., Li, J. H., Zhu, W. J., Krueger, K. E. & Wang, J. F. (1998) Abstr. Soc. Neurosci. 24, 1990.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190133497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190133497
References
- 1.Craig A M, Blackstone C D, Huganir R L, Banker G. Proc Natl Acad Sci USA. 1994;91:12373–12377. doi: 10.1073/pnas.91.26.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whatley V J, Harris R A. Int Rev Neurobiol. 1996;39:113–143. doi: 10.1016/s0074-7742(08)60665-0. [DOI] [PubMed] [Google Scholar]
- 3.Whatley V J, Mihic S J, Allan A M, McQuilkin S J, Harris R A. J Biol Chem. 1994;269:19546–19552. [PubMed] [Google Scholar]
- 4.Crestani F, Lorez M, Baer K, Essrich C, Benke D, Laurent J P, Belzung C, Fritschy J M, Luscher B, Mohler H. Nat Neurosci. 1999;2:833–839. doi: 10.1038/12207. [DOI] [PubMed] [Google Scholar]
- 5.Sassoe-Pognetto M, Kirsch J, Grunert U, Greferath U, Fritschy J M, Mohler H, Betz H. J Comp Neurol. 1995;357:1–14. doi: 10.1002/cne.903570102. [DOI] [PubMed] [Google Scholar]
- 6.Todd A J, Watt C, Spike R C, Sieghart W. J Neurosci. 1996;16:974–982. doi: 10.1523/JNEUROSCI.16-03-00974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essrich C, Fritschy J M, Lorez M, Benson J, Luscher B. Nat Neurosci. 1998;1:563–571. doi: 10.1038/2798. [DOI] [PubMed] [Google Scholar]
- 8.Meyer G, Kirsch J, Betz H, Langosch D. Neuron. 1995;15:563–572. doi: 10.1016/0896-6273(95)90145-0. [DOI] [PubMed] [Google Scholar]
- 9.Betz H. Nat Neurosci. 1998;1:541–543. doi: 10.1038/2777. [DOI] [PubMed] [Google Scholar]
- 10.Gautam M, Noakes P, Mudd J, Nichol M, Chu G, Sanes J, Merlie J. Nature (London) 1995;377:232–236. doi: 10.1038/377232a0. [DOI] [PubMed] [Google Scholar]
- 11.Yang S H, Armson P F, Cha J, Phillips W D. Mol Cell Neurosci. 1997;8:430–438. doi: 10.1006/mcne.1997.0597. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Bedford F K, Brandon N J, Moss S J, Olsen R W. Nature (London) 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- 13.Wang H B, Olsen R W. J Neurochem. 2000;75:644–655. doi: 10.1046/j.1471-4159.2000.0750644.x. [DOI] [PubMed] [Google Scholar]
- 14.Bueno O F, Robinson L C, Alvarez-Hernandez X, Leidenheimer N J. J Biol Chem. 1998;273:32595–32601. doi: 10.1074/jbc.273.49.32595. [DOI] [PubMed] [Google Scholar]
- 15.Jones M V, Westbrook G L. Neuron. 1995;15:181–191. doi: 10.1016/0896-6273(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, Huang L-Y M. Neuron. 1991;6:777–784. doi: 10.1016/0896-6273(91)90174-x. [DOI] [PubMed] [Google Scholar]
- 17.Haas K F, Macdonald R L. J Physiol (London) 1999;514:27–45. doi: 10.1111/j.1469-7793.1999.027af.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneda M, Farrant M, Cull-Candy S G. J Physiol (London) 1995;485:419–435. doi: 10.1113/jphysiol.1995.sp020739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brickley S G, Cull-Candy S G, Farrant M. J Physiol (London) 1996;497:753–759. doi: 10.1113/jphysiol.1996.sp021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brickley S G, Cull-Candy S G, Farrant M. J Neurosci. 1999;19:2960–2973. doi: 10.1523/JNEUROSCI.19-08-02960.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison D W, Chervin A S, Gelfand V I, Graig A M. J Neurosci. 2000;20:4545–4554. doi: 10.1523/JNEUROSCI.20-12-04545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lester R A, Clements J D, Westbrook G L, Jahr C E. Nature (London) 1990;346:565–567. doi: 10.1038/346565a0. [DOI] [PubMed] [Google Scholar]
- 23.Diamond J S, Jahr C E. Neuron. 1995;15:1097–1107. doi: 10.1016/0896-6273(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 24.Jones M V, Westbrook G L. J Neurosci. 1997;17:7626–7633. doi: 10.1523/JNEUROSCI.17-20-07626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Huang L-Y M. Nature (London) 1992;356:521–523. doi: 10.1038/356521a0. [DOI] [PubMed] [Google Scholar]
- 26.De Koninck Y, Mody I. J Neurophysiol. 1994;71:1318–1335. doi: 10.1152/jn.1994.71.4.1318. [DOI] [PubMed] [Google Scholar]
- 27.Benke T A, Lüthi A, Isaac J T, Collingridge G L. Nature (London) 1998;393:793–797. doi: 10.1038/31709. [DOI] [PubMed] [Google Scholar]
- 28.Westphal R S, Tavalin S J, Lin J W, Alto N M, Fraser I D, Langeberg L K, Sheng M, Scott J D. Science. 1999;285:93–96. doi: 10.1126/science.285.5424.93. [DOI] [PubMed] [Google Scholar]
- 29.Funk G D, Smith J C, Feldman J L. J Neurosci. 1995;15:4046–4056. doi: 10.1523/JNEUROSCI.15-05-04046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raines D E, Rankin S E, Miller K W. Anesthesiology. 1995;82:276–287. doi: 10.1097/00000542-199501000-00033. [DOI] [PubMed] [Google Scholar]
- 31.Huang C S, Ma J Y, Marszalec W, Narahashi T. Neuropharmacology. 1996;35:1251–1261. doi: 10.1016/s0028-3908(96)00074-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhu W J, Vicini S. J Neurosci. 1997;17:4022–4031. doi: 10.1523/JNEUROSCI.17-11-04022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen R W. Toxicol Lett. 1998;100–101:193–201. doi: 10.1016/s0378-4274(98)00185-4. [DOI] [PubMed] [Google Scholar]
- 34.Zorumski C F, Mennerick S J, Covey D F. Synapse. 1998;29:162–171. doi: 10.1002/(SICI)1098-2396(199806)29:2<162::AID-SYN7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 35.Angelotti T P, Macdonald R L. J Neurosci. 1993;13:1429–1440. doi: 10.1523/JNEUROSCI.13-04-01429.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huganir R L, Greengard P. Neuron. 1990;5:555–567. doi: 10.1016/0896-6273(90)90211-w. [DOI] [PubMed] [Google Scholar]
- 37.Sigel E. J Rec Sig Trans Res. 1995;15:325–332. doi: 10.3109/10799899509045224. [DOI] [PubMed] [Google Scholar]
- 38.Smart T G. Curr Opin Neurobiol. 1997;7:358–367. doi: 10.1016/s0959-4388(97)80063-3. [DOI] [PubMed] [Google Scholar]
- 39.McDonald B J, Amato A, Connolly C N, Benke D, Moss S J, Smart T G. Nat Neurosci. 1998;1:23–28. doi: 10.1038/223. [DOI] [PubMed] [Google Scholar]
- 40.Nusser Z, Sieghart W, Somogyi P. J Neurosci. 1998;18:1693–1703. doi: 10.1523/JNEUROSCI.18-05-01693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wall M J, Usowicz M M. Eur J Neurosci. 1997;9:533–548. doi: 10.1111/j.1460-9568.1997.tb01630.x. [DOI] [PubMed] [Google Scholar]
- 42.Rossi D J, Hamann M. Neuron. 1998;20:783–795. doi: 10.1016/s0896-6273(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 43.Tia S, Wang J F, Kotchabhakdi N, Vicini S. Neuropharmacology. 1996;35:1375–1382. doi: 10.1016/s0028-3908(96)00018-4. [DOI] [PubMed] [Google Scholar]
- 44.McClellan A M L, Twyman R E. J Physiol (London) 1999;515:711–727. doi: 10.1111/j.1469-7793.1999.711ab.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maconochie D J, Knight D E. J Physiol (London) 1992;454:129–153. doi: 10.1113/jphysiol.1992.sp019257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salamone F N, Zhou M, Auerbach A. J Physiol (London) 1999;516:315–330. doi: 10.1111/j.1469-7793.1999.0315v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall Z W, Sanes J R. Neuron. 1993;11:99–126. [Google Scholar]