Abstract
The induction of the dehydration-responsive Arabidopsis gene, rd29B, is mediated mainly by abscisic acid (ABA). Promoter analysis of rd29B indicated that two ABA-responsive elements (ABREs) are required for the dehydration-responsive expression of rd29B as cis-acting elements. Three cDNAs encoding basic leucine zipper (bZIP)-type ABRE-binding proteins were isolated by using the yeast one-hybrid system and were designated AREB1, AREB2, and AREB3 (ABA-responsive element binding protein). Transcription of the AREB1 and AREB2 genes is up-regulated by drought, NaCl, and ABA treatment in vegetative tissues. In a transient transactivation experiment using Arabidopsis leaf protoplasts, both the AREB1 and AREB2 proteins activated transcription of a reporter gene driven by ABRE. AREB1 and AREB2 required ABA for their activation, because their transactivation activities were repressed in aba2 and abi1 mutants and enhanced in an era1 mutant. Activation of AREBs by ABA was suppressed by protein kinase inhibitors. These results suggest that both AREB1 and AREB2 function as transcriptional activators in the ABA-inducible expression of rd29B, and further that ABA-dependent posttranscriptional activation of AREB1 and AREB2, probably by phosphorylation, is necessary for their maximum activation by ABA. Using cultured Arabidopsis cells, we demonstrated that a specific ABA-activated protein kinase of 42-kDa phosphorylated conserved N-terminal regions in the AREB proteins.
The phytohormone abscisic acid (ABA) plays important roles in the adaptation of vegetative tissues to abiotic environmental stresses such as drought and high salinity, as well as in seed maturation and dormancy. ABA promotes stomatal closure in guard cells, mediating by solute efflux, and regulates the expression of many genes, the products of which may function in dehydration tolerance in both vegetative tissues and seeds (1–3). Many ABA-inducible genes contain a conserved, ABA-responsive, cis-acting element named ABRE (ABA-responsive element; PyACGTGGC) in their promoter regions (1–3). ABRE functions as a cis-acting DNA element involved in ABA-regulated gene expression. ABREs were first identified in the wheat Em gene, which functions mainly in seed during late embryogenesis (4), and in the rice rab16 gene, which is expressed in both dehydrated vegetative tissues and maturating seeds (5). A cDNA for the ABRE-binding protein EmBP-1 was first shown to encode a basic leucine zipper (bZIP) protein containing a basic DNA-binding domain linked to a leucine zipper domain (4). However, a single copy of ABRE is not sufficient for ABA-responsive transcription. Furthermore, the G-box resembles the ABRE motif and functions in the regulation of plant genes in a variety of environmental conditions, such as red light, UV light, anaerobiosis, and wounding (6). G-box-binding proteins also contain a bZIP motif (6). Nucleotides around the ACGT core motif are important for determining the binding specificity of bZIP proteins. Furthermore, a coupling element is required to specify the function of ABRE as an ABRE. ABRE and a coupling element constitute an ABA-responsive complex in the regulation of the wheat HVA22 gene (7). Most of the known coupling elements have similarity with ABREs and contain an A/GCGT motif (8). Several genes for bZIP that bind to ABREs in vitro or are inducible by ABA have been isolated (1–3). However, none of their functions have been clearly determined in ABA signaling, because of a lack of genetic analysis in these studies.
The regulatory machinery in ABA-responsive gene expression in vegetative tissues is thought to be different from that in seeds. Mutations that alter the sensitivity to ABA in seeds have been described in maize and Arabidopsis (9, 10). The maize VP1 and Arabidopsis ABI3 genes play key roles in ABA-dependent seed maturation, and both genes encode seed-specific, homologous transcription factors that mediate ABA-regulated gene expression in seeds (10, 11). The existence of VP1 homologues in vegetative tissues has not been reported thus far. In addition, some reports concerning the promoter analysis of the ABA-responsive genes showed that different cis-acting elements of the promoters regulate the expression of the genes by ABA between vegetative tissues and seeds (12, 13). These suggest that different members of transcription factors regulate the ABRE-dependent gene expression in response to dehydration stress.
On the other hand, the involvement of protein phosphatase 2C and farnesyltransferase in ABA signaling were revealed by cloning of the ABI1, ABI2, and ERA1 genes in Arabidopsis (14–17). The abi1 and abi2 mutants show decreased sensitivities to ABA inhibition of seedling growth and defects in molecular responses to applied ABA in vegetative tissues (9). The era1 mutant shows prolonged seed dormancy owing to an enhanced response to ABA (17). The ERA1, ABI1 and ABI2 genes are expressed in vegetative tissues and are thought to be essential for negative regulation of ABA signaling during vegetative growth (14–17). To understand the ABA signaling pathway during dehydration stress, it is critical to identify the transcription factors that function downstream of ABI1, ABI2, and ERA1 proteins in ABA signaling in Arabidopsis.
Two genes, rd29A and rd29B, which are located close to each other on the Arabidopsis genome, are differentially regulated by drought stress and ABA treatment (18). The promoter region of rd29A contains at least two cis-acting elements, DRE (dehydration responsive element) and ABRE, involved in ABA-independent and ABA-responsive gene expression under drought stress. In contrast, the rd29B promoter contains ABRE but not DRE (18). rd29B mRNA did not accumulate during dehydration stress in the ABA-deficient aba1 mutant (19), and its level was very low in the ABA-insensitive abi1 mutant of Arabidopsis (9), indicating that the drought-inducible expression of rd29B is mainly controlled by ABA. We analyzed the rd29B promoter in transgenic plants and found that two ABREs function as cis-regulatory elements in ABA-dependent expression of rd29B. We cloned three different cDNAs encoding ABRE-binding proteins (AREB1, AREB2, and AREB3) of Arabidopsis by using the yeast one-hybrid screening method. Each AREB protein contained a single bZIP-type DNA-binding domain, and genes encoding the AREB1 and AREB2 proteins were up-regulated by ABA. We analyzed the function of the AREB1 and AREB2 proteins as trans-acting factors by using transient expression in leaf protoplasts prepared from wild-type or ABA-related mutants. We also analyzed the ABA-specific phosphorylation of the AREB1 and AREB2 proteins by using cultured Arabidopsis cells. We discuss the functions of the AREB proteins in the ABA-dependent signaling pathways in vegetative tissues under dehydration stress conditions.
Materials and Methods
Plant Materials and Stress Treatments.
Plants (Arabidopsis thaliana ecotype Columbia) were grown on germination medium agar plates for 3 weeks as described (18). Various stress treatments and treatment with ABA were applied as described (18). T87 cells, derived from Arabidopsis, were grown and treated as described (20).
Base Substitution Analysis of the 77-bp Region of the rd29B Promoter.
Six 77-bp fragments with base substitutions (M1–M6) between positions −169 and −93 of the rd29B promoter with HindIII sites at both ends were prepared by PCR (Fig. 1). The products were cloned into the HindIII site of pBluescript II SK− (Stratagene), and the resulting plasmids were confirmed by sequencing. The tandemly repeated, dimeric, 77-bp fragments with or without base substitution were ligated to the HindIII site of the −51 rd29B minimal TATA promoter-GUS (β-glucuronidase) fusion construct (18). Confirmation of the fusion constructs, transformation of Nicotiana tabacum cv SR1, and measurement of GUS activity were performed as described (18).
Figure 1.
Base substitution analysis of the 77-bp region of the rd29B promoter involved in ABA- and dehydration-responsive expression in transgenic tobacco. GUS activity before and after treatment was measured in 20 independent transformants for each construct and is shown as average values. MU, 4-methyl-umbelliferone. (A) Upper-strand sequences of the 77-bp fragment (wild type) and its mutated sequences (M1-M6). The monomer (x1) or tandemly repeated dimer (x2) of each 77-bp fragment containing different mutations was ligated to the −51 rd29B minimal TATA promoter-GUS construct, respectively. Dashes indicate the wild-type sequence. (B) Effect of base substitutions in the ABRE and MYB recognition sites for dehydration-responsive expression of the rd29B. Half of the leaf from T1 tobacco plant was used immediately for the assay of GUS activity (control), and the other half was dehydrated for 24 h (dry). (C) Effect of ABA treatment on the induction of the GUS reporter gene driven by the 77-bp fragment. T2 tobacco seedlings were used immediately for the assay of GUS activity (control), transferred from agar plates for hydroponic growth in water (H2O) or 100 μM ABA (ABA) solution, or dehydrated for 24 h (dry).
Yeast One-Hybrid Screening of Arabidopsis cDNA Libraries.
Construction of reporter plasmids and selection of the yeast reporter strain were performed as described (21). Arabidopsis cDNA libraries were prepared from dehydrated and unstressed Arabidopsis plants as described (21). We screened 1.0 × 106 yeast transformants from each library according to the manufacturer's protocol (CLONTECH Matchmaker one-hybrid system). We obtained 230 positive colonies from selective-medium plates [15 mM 3-aminotriazole (3-AT)]. Growth of these clones was examined at 30, 45, and 60 mM 3-AT. The β-galactosidase activities of the clones then were further analyzed. Finally, 29 clones, which grew normally on the 30-mM 3-AT plate and had βgalactosidase activity, were selected. The cDNA isolation, subcloning, and sequencing of these 29 clones were performed as described (21). To analyze the binding specificity of isolated cDNA clones, four tandemly repeated copies of the mutated 77-bp fragment containing two ABRE sequences replaced with AATCAAT (Fig. 1A) were used for the construction of the reporter plasmids.
RNA Gel Blot Analyses.
RNA gel blot hybridization was performed as described (18).
Transactivation Experiment with Protoplasts.
Effector plasmids used in the transient transactivation experiment were constructed with DNA fragments containing the AREB1, AREB2, or GBF3 coding regions, which were cloned into NotI sites of the plant expression vector pBI35SΩ. The pBI35SΩ vector was constructed as described (20). To construct a reporter plasmid, the 35S promoter of pBI221 was replaced with the rd29B minimal TATA promoter, and the 77-bp fragment of the rd29B promoter then was ligated into the HindIII site located upstream from the rd29B minimal TATA promoter. A transactivation experiment with Arabidopsis mesophyll protoplasts was performed as described (20).
In-Gel Kinase Assay.
The ABA-dependent phosphorylation of the AREB proteins was analyzed by an in-gel protein kinase assay as described (22). GST-fused AREB proteins expressed in Escherichia coli were used as substrates in 10% SDS-polyacrylamide gel (50 μg/ml). Arabidopsis T87 cells were incubated in the presence or absence of 50 μM ABA for various periods and ground with mortar and pestle in liquid nitrogen; then proteins were extracted in 1 ml/g (fresh weight) extraction buffer as described (23). Cell extract proteins (10 μg) were denatured and electrophoresed in 10% SDS-polyacrylamide gel containing the GST-fused AREB proteins, and then kinase activity was assessed (22).
Reverse Transcription–PCR.
Reverse transcription–PCR was performed according to the manufacturer's protocol (Amersham Pharmacia).
Results
Identification of Cis-Acting Elements Involved in Dehydration-Responsive Expression of rd29B.
A promoter analysis with rd29B-GUS fusion genes indicated that the 77-bp region between −169 and −93 contains cis-acting elements that are involved in the dehydration- and ABA-responsive expression of rd29B (Fig. 1). The 77-bp region contains two closely located ABRE motifs (ACGTGGC and TACGTGTC) and one putative recognition site (CAACTG) for MYB-related transcription factors. First, we determined which motifs are the cis-acting elements involved in the dehydration-induced transcription of rd29B. We prepared the 77-bp fragments with or without base substitutions in two ABREs and the MYB recognition sites (Fig. 1A). The monomer or tandemly repeated dimer forms of these fragments were fused upstream of the −51 rd29B minimum promoter-GUS fusion construct and introduced into a tobacco chromosome by Agrobacterium-mediated transformation.
We analyzed 20 independent transgenic tobacco plants for the expression of each fusion gene (Fig. 1). The monomer form of the wild-type 77-bp fragment induced a 4.9-times increase in GUS (Fig. 1B). The dimer form of the wild-type 77-bp sequence induced a 33.9-times increase in GUS activity after dehydration treatment (Fig. 1B, wild type). The dimer forms of the 77-bp fragment with base substitutions in both ABREs did not function in the dehydration-induced GUS expression (Fig. 1B, M1). The 77-bp fragment with base substitution in either ABRE showed considerably reduced GUS induction (Fig. 1B, M2 and M3). In contrast, the 77-bp fragment with base substitutions in the MYB recognition site (Fig. 1B, M4) or those outside the ABRE and MYB sites (Fig. 1B, M5 and M6) responded to dehydration stress at the same level as or higher than the wild-type 77-bp fragment did (Fig. 1B). These results demonstrate that at least two ABREs function as positive, cis-acting elements in dehydration-responsive expression of rd29B. As tobacco plants could not absorb ABA easily we analyzed ABA-responsive expression of the 77-bp promoter region by using T2 tobacco seedlings of the transgenics. The level of GUS induction by ABA was similar to that by dehydration in the seedling (Fig. 1C).
Isolation of cDNAs Encoding DNA-Binding Proteins that Recognize ABRE in the 77-bp DNA Fragment of the rd29B Promoter.
To isolate cDNAs encoding DNA-binding proteins that interact with the ABRE motif, we carried out yeast one-hybrid screening. We first constructed a parental yeast strain carrying as reporter genes integrated copies of HIS3 and lacZ with four-time tandemly repeated, 77-bp DNA fragments of the rd29B promoter. The resulting yeast cells then were separately transformed with two expression libraries prepared from Arabidopsis rosette plants that had been dehydrated for 2 h or not dehydrated. The cDNA fragments were fused to the transcriptional activation domain of yeast GAL4. We isolated 23 3-AT-resistant clones from a library prepared from dehydrated plants and six clones from a library prepared from undehydrated plants. All of the isolated cDNA clones induced lacZ activity and formed blue colonies on filter papers containing 5-bromo-4-chloro-3-indolyl β-d-galactoside. The cDNA fragments of the isolated plasmids were analyzed by restriction enzyme digestion and DNA sequencing, which led to the classification of these 29 cDNA clones into five distinct cDNA groups. Two were the same as previously identified clones GBF1 and GBF3 (24), and three were novel clones that were designated AREB1, AREB2, and AREB3 (Fig. 2A).
Figure 2.
Isolation of cDNAs encoding ABRE-binding proteins by the yeast one-hybrid system. (A) Confirmation of five isolated cDNAs encoding ABRE-binding proteins by the yeast one-hybrid system. Five plasmids containing insert DNA from AREB1, AREB2, AREB3, GBF1, and GBF3 were retransformed into yeast strains carrying the reporter genes HIS3 and lacZ under the control of the 77-bp fragment containing two ABREs. The transformants were examined for growth in the presence of 3-AT and β-galactosidase (β-gal) activity. (B) Binding specificity of proteins encoded by isolated cDNAs. Five plasmids were used for transformation into yeast carrying the reporter genes under the control of the 77-bp fragment containing mutated ABREs. (C) Activation of reporter genes in yeast by proteins encoded by isolated cDNAs. The insert DNA fragments of the five isolated cDNA clones were cloned into the yeast expression vector YepGAP and used for transformation into yeast carrying the reporter genes under the control of the 77-bp fragment containing two ABREs.
When the five isolated plasmids were transformed into yeast strains carrying the reporter genes fused to the 77-bp DNA fragment with base substitution in the ABRE sequences, the yeast strains neither grew on medium lacking histidine in the presence of 3-AT nor induced lacZ activity (Fig. 2B). To select cDNAs that encode transcriptional activators in the five isolated clones, the insert cDNA fragments were cloned into the yeast expression vector YepGAP. Plasmids containing each insert DNA were transformed into yeast strains carrying the reporter genes, which had been fused to the 77-bp DNA. Yeast cells carrying the plasmid containing the cDNAs of AREB1, AREB2, and AREB3 grew on a medium lacking histidine in the presence of 3-AT, but yeast cells carrying the plasmid containing the cDNAs of GBF1 and GBF3 did not (Fig. 2C). All of the 3-AT-resistant yeast strains also formed blue colonies. These data indicate that cDNAs of AREB1, AREB2, and AREB3 encode polypeptides that specifically bind to the ABRE sequence and activate the transcription of the reporter genes in yeast. These three clones were analyzed further.
Structural Analysis of the AREB cDNAs.
To examine the structures of the AREB1, AREB2, and AREB3 cDNA clones, we sequenced inserted DNA fragments of 1.6, 1.6, and 0.7 kb, respectively. The AREB1 cDNA contained a single ORF of 416 aa and encoded a putative protein with a predicted molecular mass of 44.2 kDa (Fig. 3). The AREB2 cDNA contained an ORF of 431 aa and encoded a putative protein with a predicted molecular mass of 46.5 kDa. Analysis of the AREB3 cDNA revealed that the 0.7-kb DNA insert was not full length. A full-length cDNA library was screened with the AREB3 insert (25), and then a cDNA clone containing the entire coding region of this gene was isolated. The full-length AREB3 cDNA contained a single ORF of 297 aa and encoded a putative protein with a predicted molecular mass of 32.4 kDa.
Figure 3.
Comparison of deduced amino acid sequences of the AREB 1, AREB2, and AREB3 proteins and sunflower DPBF1 (21). Symbols denote identical (*) and conserved (⋅) amino acid residues in the four sequences. The boxes represent conserved regions, and dashes indicate gaps introduced to maximize alignment. Inverted characters indicates consensus sequences for various kinases (R/KXXS/T for CDPK, S/TXXD/E for CK II, S/TXK/R for PKC, K/RXXXS/T for cGMP-dependent protein kinase) appeared within the conserved regions.
Homology search of DNA and protein databases indicated that the AREB1, AREB2, and AREB3 proteins have a bZIP-type DNA-binding domain (Fig. 3). Four or three heptad repeats of leucine or isoleucine exist near the C terminus in the three AREB proteins. The basic domains of the AREB proteins are nearly identical to each other, which suggests that these DNA-binding proteins recognize the same target sequence. The three AREB proteins contain the three conserved sequences in their N-terminus half regions and a short amino acid sequence near their C terminus. Two of the conserved amino acid sequences of the AREB proteins showed significant sequence identity in their N-terminus regions to the sunflower DPBF1 protein, which binds to ABA-responsive and embryo-specification elements in the carrot Dc3 promoter (ref. 26; Fig. 3).
Expression of the AREB Genes.
The expression patterns of the AREB1, AREB2, and AREB3 genes were analyzed by RNA gel blot hybridization to compare them with that of the rd29B gene (Fig. 4). The rd29B gene was induced within 2 h after dehydration, high-salt treatment, or exogenous ABA treatment. The AREB1 gene expression was induced within 2 h after dehydration began, and the level of the AREB1 mRNA increased over 24 h. The induction of AREB2 was detected within 24 h after dehydration stress. The induction of the AREB1 and AREB2 mRNAs also was observed within 2 h after high-salt or exogenous ABA treatment. The AREB3 mRNA was not detected in any stressed or unstressed plants (data not shown). These data indicate that AREB1 and AREB2 gene products most likely function as transcription factors involved in the induction of rd29B. The weak expression of both AREB1 and AREB2 was detected in untreated plants. Tissue-specific expression also was analyzed and found that both AREB1 and AREB2 were expressed in roots and leaves but not in seeds (data not shown).
Figure 4.
(A) Expression of the AREB1, AREB2, and rd29B genes in response to dehydration, low temperature, high salt, or ABA. Each lane was loaded with 40 μg of total RNA from 3-week-old Arabidopsis plants that had been dehydrated (Dry), transferred to 4°C (Cold), transferred to hydroponic growth in 250 mM NaCl (NaCl), transferred to hydroponic growth in 100 μM ABA (ABA), or transferred to water (H2O), as described in Materials and Methods. The number above each lane indicates the number of minutes or hours after the initiation of the treatment. rRNAs blotted on the membrane were visualized by staining with methylene blue.
The AREB1 and AREB2 Proteins Transactivate the rd29B Promoter-GUS Fusion Gene in Leaf Protoplasts.
The rd29B gene expression under various stress conditions was analyzed by RNA gel blot hybridization in wild-type or ABA-related mutants (Fig. 5A). The rd29B gene was induced by dehydration, high-salt treatment, or exogenous ABA treatment in wild-type plants (Columbia and Landsberg) and in the abi3 mutant. However, the rd29B mRNA did not accumulate during dehydration or high-salt stress in the aba1 mutant, and its level was very low in the abi1 mutant. The rd29B gene was not induced by ABA in the abi1 mutant. These observations suggest that the dehydration- and high-salt-responsive expression of rd29B is regulated mainly by ABA, and that this ABA-dependent expression is mediated by the ABI1 protein but not by ABI3. Previously, Nordin et al. (27) also have shown similar results analyzing the expression of lti65/rd29B.
Figure 5.
Transactivation of the rd29B promoter-GUS fusion gene by AREB1 and AREB2 proteins using Arabidopsis protoplasts. (A) Expression of rd29B in response to dehydration, low temperature, high salt, or ABA in wild type (Columbia & Landsberg) or ABA-related mutants (aba2, abi1, and abi3). Plants were untreated (C), dehydrated for 10 h (D), or treated with water (H), 250 mM NaCl (N) or 100 μM ABA (A) for 5 h. (B) Schematic diagram of the effector and reporter constructs used in cotransfection experiments. The effector constructs contain the CaMV 35S promoter and TMV Ω sequence fused to AREB1, AREB2, or GBF3 cDNAs. The reporter constructs contain the 77-bp fragments of the rd29B promoter connected tandemly five times (×5) or single (×1). The promoters were fused to the −51 rd29B minimal TATA promoter-GUS construct. (C) Transactivation of the rd29B promoter-GUS fusion gene by AREB1, AREB2, and GBF3 proteins. The reporter gene driven by the 77-bp fragments tandemly repeated five times was transfected with each effector plasmid or the vector as control treatments. Transactivation experiments using protoplasts prepared from wild type or ABA-mutant (abi1, aba2, and era1) Arabidopsis leaves. To normalize for transfection efficiency, the CaMV 35S promoter-luciferase (LUC) plasmid was cotransfected in each experiment. Bars indicate the standard error of three replicates. Ratios indicate the multiples of expression compared with the value obtained with the pBI35SΩ vector, and ratio in parentheses indicates the multiples of expression compared with value obtained with the wild type transformed with pBI35SΩ vector. (D) Transactivation using the reporter construct containing the single 77-bp fragment.
To determine whether the AREB1 and AREB2 proteins are capable of transactivating ABRE-dependent transcription in plant cells, we performed transactivation experiments using protoplasts prepared from Arabidopsis leaves. Protoplasts were cotransfected with a GUS reporter gene fused to the five tandem copies of the 77-bp fragment containing two ABRE motifs and an effector plasmid (Fig. 5B). The effector plasmid consisted of the CaMV 35S promoter fused to the AREB1 or AREB2 cDNAs. Coexpression of the AREB1 or AREB2 proteins in protoplasts derived from wild-type Arabidopsis plants transactivated the expression of the GUS reporter gene, but that of GBF3 protein did not (Fig. 5C). These results suggest that AREB1 and AREB2 function as transcription activators involved in the ABRE-dependent expression of rd29B. Addition of ABA into protoplasts further activated the expression of the GUS reporter gene (Fig. 5C). To investigate activation of a native ABA-inducible promoter by AREB1 and AREB2, leaf protoplasts were cotransfected with a GUS reporter gene fused to the monomer of the 77-bp fragment of the rd29B promoter (Fig. 5B). We obtained similar activation of GUS expression by using the monomer form of the promoter region of rd29B (Fig. 5D).
We isolated leaf protoplasts from several ABA-related mutants. In the aba2 mutant background, transactivation of AREB1 and AREB2 was decreased to the basal level but was strongly increased by the addition of ABA (Fig. 5C). Transactivation of the AREB proteins was significantly decreased in protoplasts derived from the abi1 mutant (Fig. 5C). We obtained similar results by using protoplasts prepared from the abi2 mutant (data not shown). By contrast, the transactivation of the AREB proteins was strongly activated by ABA in the era1 mutant (Fig. 5C). These results suggest that overproduction of the AREB1 and AREB2 proteins alone is not sufficient for the expression of rd29B. Activation of AREB1 by ABA was repressed by the addition of a protein kinase inhibitor, staurosporine (Fig. 5C). We obtained similar results by adding K252a (data not shown). Modification of the AREB proteins, such as phosphorylation in response to ABA, may be necessary for their activation.
ABA-Specific Phosphorylation of AREB1 and AREB2 Proteins.
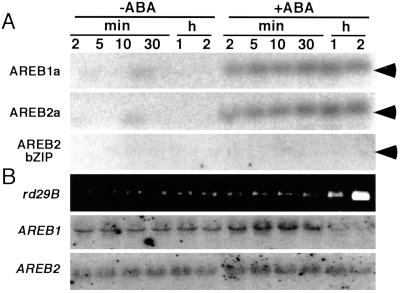
ABA-dependent phosphorylation of the AREB proteins was examined by using an in-gel protein kinase assay. The protein kinase activities were measured in polyacrylamide gels containing the recombinant AREB protein by using cell extracts from ABA-treated Arabidopsis T87 culture cells. We detected ABA-dependent phosphorylation of the AREB proteins by 42-kDa protein kinases when we used a recombinant polypeptide of the AREB proteins containing the first conserved sequences in their N-terminal regions as substrates (Fig. 6A, AREB1a and AREB2a). Polypeptides containing the second and third conserved regions also were phosphorylated, but those of unconserved regions (data not shown) and the DNA-binding domain (Fig. 6A) were not. This protein kinase activity was activated by ABA within 2 min and reached a maximum level at 1 h. The rd29B mRNA accumulated in response to ABA in Arabidopsis culture cells, whereas the expression of the AREB1 and AREB2 genes was constant during 2 h (Fig. 6B).
Figure 6.
ABA-dependent phosphorylation of recombinant AREB proteins by in-gel kinase activity assay. (A) T87 cell extracts treated with or without 100 μM ABA treatment were resolved on 10% polyacrylamide gel containing recombinant AREB proteins (AREB1a: 73G-131Q; AREB2a: 84S-133D; AREB2 bZIP domain: 335Y-401V). The protein kinase activities were analyzed as described in Materials and Methods. (B) Accumulation of the rd29B, AREB1, and AREB2 mRNAs in T87 culture cells. The rd29B mRNA was detected by reverse transcription–PCR, and the AREB1 and AREB2 mRNAs were detected by RNA gel blot analysis.
Discussion
The base substitution experiments demonstrated that at least two ABREs in the 77-bp region of the rd29B promoter are involved in dehydration- and ABA-responsive expression (Fig. 1). It has been shown that polymerized copies of ABREs can confer ABA responsiveness to a minimal promoter, whereas a single copy of ABRE was not sufficient for the full ABA response (28). In the case of the wheat Em promoter, both the Em1a and Em1b ABRE motifs contribute to the promoter activity (4). In the barley HVA1 promoter, the typical ABRE motifs and the coupling element CE3 directly upstream of ABRE are shown to be sufficient for ABA induction of gene expression. The CE3 sequence (ACGCGTGTCCTC) is similar to a typical ABRE (ACGTGG/TC; ref. 29), and other coupling elements also contain an A/GCGT core sequence (8). The existence of more than two ABREs at appropriate positions in the promoter regions seems to be sufficient for the ABA-inducible transcription.
We identified three distinct cDNAs (AREB1, AREB2, and AREB3)-encoding DNA-binding proteins that specifically interact with ABREs involved in dehydration- and ABA-responsive gene expression of rd29B. These three AREB genes encode novel bZIP-type proteins. The AREB1 and AREB2 genes were shown to be expressed weakly in unstressed plants and to be induced by dehydration, high-salt treatment, and endogenous ABA treatment, whereas the AREB3 mRNA was not detected in either stressed or unstressed plants. Both the AREB1 and AREB2 proteins functioned as transcriptional activators for ABRE-dependent transcription, not only in yeast cells (Fig. 2), but also in Arabidopsis leaf protoplasts (Fig. 5). Other bZIP factors, GBF-1 and GBF-3, also were cloned with the yeast one-hybrid screening. However, neither GBF protein transactivated the reporter genes fused to the rd29B promoter in yeast (Fig. 2). The GBF-3 protein did not function as a transcriptional activator for the ABA-inducible expression of rd29B in Arabidopsis leaf protoplasts (Fig. 5C). These results strongly suggest that the AREB1 and AREB2 proteins are specific bZIP transcription factors that are involved in ABRE-dependent expression of rd29B.
The expression analysis of rd29B in ABA-related mutants indicated that the dehydration- and high-salt-responsive gene expression of rd29B is mediated mainly by ABA (Fig. 5A). We carried out transactivation experiments using leaf protoplasts prepared from ABA-related Arabidopsis mutants. The overexpression of the AREB proteins was not sufficient for the induction of rd29B, and ABA treatment was required for the activation of AREBs (Fig. 5C). Posttranscriptional activation of the AREB proteins in response to ABA may be necessary for their activities. Transactivation by AREBs was significantly decreased in the ABA-insensitive abi1 and ABA-deficient aba2 mutants, but was increased extremely in the ABA-hypersensitive era1 mutant. These results suggest that the ABI1, ABI2, and ERA1 proteins are involved in the upstream signal transduction pathway of the ABRE-dependent gene expression of rd29B.
Protein phosphorylation of the AREB proteins was suggested to be involved in their activation (Fig. 5C). So we analyzed posttranscriptional activation of AREB proteins by ABA using Arabidopsis T87 cultured cells and showed that N-terminal conserved regions of the AREB1 and AREB2 proteins were rapidly phosphorylated by a 42-kDa protein kinase (Fig. 6A). The AREB proteins have three conserved sequences in their N-terminal regions and a conserved sequence in their C-terminal regions (Fig. 3). These conserved regions contain possible target sequences for Ser/Thr protein kinases (Fig. 3). Polypeptides containing the other conserved sequences also were phosphorylated, whereas other polypeptides were not (data not shown). Phosphorylation of these conserved regions may be involved in the activation of the AREB proteins by ABA.
To identify the target sequence of the AREB1 and AREB2 proteins, the AREB1 and AREB2 DNA-binding domains were expressed as glutathione S-transferase fusion proteins in E. coli. The ability of the AREB1 and AREB2 fusion proteins to bind the wild-type or mutated sequences was examined by the gel retardation method. Both the recombinant AREB1 and AREB2 fusion proteins specifically bound to the ABRE sequence (data not shown). The ABRE sequence contains a symmetrical ACGT core element, and most plant bZIP proteins recognize sequences that have this element. Some exceptions are reported, such as maize Opaque-2, which binds to the TGACTCA element and several other sequences with a CATG core (30). The consensus binding site of the sunflower Dc3 promoter-binding factor (DPBF), ACACNNG, differs from that of the typical bZIP proteins. The sequence similarity between the AREB and DPBF proteins is high among bZIP proteins. However, the DNA-binding specificity of these proteins and the tissue-specific expression of these genes are quite different. The expression of the DPBF genes is seed-specific, whereas the AREB1 and AREB2 genes express only in vegetative tissues. Recently, Hobo et al. (31) reported that a rice bZIP transcription factor, TRAB1, has high sequence similarity with the DPBF proteins and interacts with VP1. These genes may function in VP1-dependent gene expression that controls maturation and dormancy in plant embryos. These results suggest that transcriptional regulation in ABA-responsive gene expression in vegetative tissues is different from that in seeds. Further analysis of the function of the AREB1 and AREB2 proteins will lead us to an understanding of the linkage between ABA-dependent gene expression and upstream signaling molecules, including ABI1/2 and ERA1, in vegetative tissues.
Acknowledgments
We thank Ms. Atsuko Iuchi, Ms. Ekuko Ohgawara, and Ms. Mie Yamamoto for their excellent technical assistance. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences.
Abbreviations
- ABA
abscisic acid
- ABRE
ABA-responsive element
- AREB
ABA-responsive element binding protein
- bZIP
basic leucine zipper
- GUS
β-glucuronidase
- 3-AT
3-aminotriazole
- DPBF
Dc3 promoter-binding factor
Note Added in Proof.
While our manuscript was under review, cDNAs related or identical to our AREB1 and AREB2 were reported (32). The ABI5 gene involved in ABA-insensitive phenotype was shown to encode AREB-related transcription factor (33).
Footnotes
Data deposition: The nucleotide sequences of the AREB1, AREB2, and AREB3 cDNAs reported in this paper have been deposited in the GenBank database (accession nos. AB017160, AB017161, and AB017162, respectively).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190309197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190309197
References
- 1.Bonetta D, McCourt P. Trends Plant Sci. 1998;3:231–235. [Google Scholar]
- 2.Grill E, Himmelbach A. Curr Opin Plant Biol. 1998;1:412–418. doi: 10.1016/s1369-5266(98)80265-3. [DOI] [PubMed] [Google Scholar]
- 3.Leung J, Giraudat J. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- 4.Guiltinan M J, Marcotte W R, Jr, Quatrano R S. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 5.Mundy J, Yamaguchi-Shinozaki K, Chua N-H. Proc Natl Acad Sci USA. 1990;87:406–410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menkes A E, Schindler U, Cashmore A R. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- 7.Shen L M, Ho T-H D. Plant Cell. 1995;7:295–307. doi: 10.1105/tpc.7.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobo T, Asada M, Kowyama Y, Hattori T. Plant J. 1999;19:679–689. doi: 10.1046/j.1365-313x.1999.00565.x. [DOI] [PubMed] [Google Scholar]
- 9.Koornneef M, Reuling G, Karssen C M. Physiol Plant. 1984;61:377–383. [Google Scholar]
- 10.McCarty D R, Hattori T, Carson C B, Vasil V, Lazar M, Vasil I K. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 11.Giraudat J, Hauge B M, Valon C, Smalle J, Parcy F, Goodman H M. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasil V, Marcotte W R, Jr, Rosenkrans L, Cocciolone S M, Vasil I K, Quatrano R S, McCarty D R. Plant Cell. 1995;7:1511–1518. doi: 10.1105/tpc.7.9.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busk P K, Jensen A J, Pages M. Plant J. 1997;11:1285–1295. doi: 10.1046/j.1365-313x.1997.11061285.x. [DOI] [PubMed] [Google Scholar]
- 14.Leung J, Bouvier-Durand M, Morris P-C, Guerrier D, Chefdor F, Giraudat J. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 15.Meyer K, Leube M P, Grill E. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 16.Leung J, Merlot S, Giraudat J. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koornneef M, Jorna M L, Brinkhorst-van der Swan D L C, Karssen C M. Theor Appl Genet. 1992;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- 20.Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Kamada H, Shinozaki K. Plant J. 1993;21:279–289. doi: 10.1007/BF00019944. [DOI] [PubMed] [Google Scholar]
- 23.Usami S, Banno H, Ito Y, Nishihama R, Machida Y. Proc Natl Acad Sci USA. 1995;92:8660–8664. doi: 10.1073/pnas.92.19.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore A R. EMBO J. 1992;11:1275–1289. doi: 10.1002/j.1460-2075.1992.tb05171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K. Plant J. 1998;15:707–720. doi: 10.1046/j.1365-313x.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim S Y, Chung H-J, Thomas T L. Plant J. 1997;11:1237–1251. doi: 10.1046/j.1365-313x.1997.11061237.x. [DOI] [PubMed] [Google Scholar]
- 27.Nordin K, Vahala T, Palva T. Plant Mol Biol. 1993;21:641–653. doi: 10.1007/BF00014547. [DOI] [PubMed] [Google Scholar]
- 28.Skriver K, Olsen F L, Rogers J C, Mundy J. Proc Natl Acad Sci USA. 1991;88:7266–7270. doi: 10.1073/pnas.88.16.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Q, Zhang P H, Ho T H. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu G, Paul A L, McCarty D R, Ferl R J. Plant Cell. 1996;8:847–857. doi: 10.1105/tpc.8.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobo T, Kowyama Y, Hattori T. Proc Natl Acad Sci USA. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi H, Hong J, Ha J, Kang J, Kim S Y. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 33.Finkelstein R R, Lynch T J. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]