Abstract
Previously, we reported that the rice dwarf mutant, d1, is defective in the α subunit of the heterotrimeric G protein (Gα). In the present study, gibberellin (GA) signaling in d1 and the role of the Gα protein in the GA-signaling pathway were investigated. Compared with the wild type, GA induction of α-amylase activity in aleurone cells of d1 was greatly reduced. Relative to the wild type, the GA3-treated aleurone layer of d1 had lower expression of Ramy1A, which encodes α-amylase, and OsGAMYB, which encodes a GA-inducible transcriptional factor, and no increase in expression of Ca2 +-ATPase. However, in the presence of high GA concentrations, α-amylase induction occurred even in d1. The GA sensitivity of second leaf sheath elongation in d1 was similar to that of the wild type in terms of dose responsiveness, but the response of internode elongation to GA was much lower in d1. Furthermore, Os20ox expression was up-regulated, and the GA content was elevated in the stunted internodes of d1. All these results suggest that d1 affects a part of the GA-signaling pathway, namely the induction of α-amylase in the aleurone layer and internode elongation. In addition, a double mutant between d1 and another GA-signaling mutant, slr, revealed that SLR is epistatic to the D1, supporting that the Gα protein is involved in GA signaling. However, the data also provide evidence for the presence of an alternative GA-signaling pathway that does not involve the Gα protein. It is proposed that GA signaling via the Gα protein may be more sensitive than that of the alternative pathway, as indicated by the low GA responsiveness of this Gα-independent pathway.
Heterotrimeric G proteins are associated with the cytoplasmic face of the plasma membrane of eukaryotic cells and mediate signalings from receptors on the cell surface. The α-subunits of heterotrimeric G (Gα) proteins transduce signals from G protein-coupled receptors to effector proteins, accompanied by the GTPase-catalyzed hydrolysis of GTP. In mammals, a set of genes has been identified for each of the G protein subunits: more than 16 genes for the α subunits, 5 genes for the β subunit, and 6 genes for the γ subunit, most of which are expressed in a tissue-specific manner. Thus mammals contain multiforms of the G proteins that are probably involved in separate systems of signal transduction. Genomic and cDNA clones that encode polypeptides similar to the mammalian Gα proteins have also been isolated from various plant species, including Arabidopsis (1), tomato (2), Lotus japonicas (3), rice (4), and soybean (5). However, with the exception of soybean, which has two genes, only a single gene has been identified in each plant species. Nevertheless, it has been proposed that the plant heterotrimeric G protein is involved in various signal transduction systems, including those of several plant hormones, blue and red light-mediated responses, pathogen resistance, and pathogen-related gene expression.
By using a constitutive stimulator (Mas7) of GDP/GTP exchange by Gα proteins, Jones et al. (6) recently demonstrated that Gα proteins might be involved in gibberellin (GA) signal transduction pathway leading to α-amylase gene expression in oat aleurone protoplasts. This is the first evidence for the involvement of the Gα proteins in GA signal transduction in plants. However, because Mas7 is also known to activate other signaling molecules, more direct evidence is required to lead a definite role for plant Gα proteins in the GA signal transduction pathway. Studies with mutants in the G protein genes should be one of the most conclusive ways to determine the role of the G protein in GA signaling in higher plants.
Recently, we (7, 8) demonstrated that the rice d1 mutant is defective in the gene that encodes the Gα protein. In the present work, we performed phenotypic analyses of the d1 mutant to investigate GA signal transduction and found that GA signaling was partially impaired in the mutant. On the basis of these observations, the role of the Gα protein in GA signaling in rice is discussed.
Materials and Methods
Plant Materials and Growth Conditions.
The rice cultivar Oryza sativa L. cv. Taichung 65 (wild type) and three rice mutants, Taichung 65-Daikoku dwarf (T65d1, dwarf), Akibare-waisei dwarf (d18-AD, dwarf), and Nipponbare slender rice (slr-1, slender), were used in this study. Rice seeds were immersed in water for 2 days, grown for 1 month in a greenhouse, and then transplanted to the field. The individual internodes, including the first to fifth internodes of the wild-type plant and the first to third internodes of the d1 mutant, were collected at the stage when the first and second internodes were elongating. Tissues were frozen in liquid N2 and stored at −80°C until extraction of RNA.
GA Induction in Shoot Elongation.
To investigate the role of GA in second leaf sheath elongation, 10 rice seeds for each rot were sterilized, pretreated with or without 6.85 μM uniconazol at 30°C for 1 day, washed 4 times with sterilized water, and then imbibed for 1 more day. The seeds were placed on agar containing various concentrations of GA3 and incubated at 30°C under continuous light. After 6 days of incubation, the lengths of the second leaf sheaths were measured.
To measure internode elongation in response to GA, 10 rice seeds were sown in a pot of soil and grown for 2 weeks in a greenhouse under 12-hr light and 12-hr dark cycles at 30°C. The seedlings were then transplanted to soil containing various concentrations of GA3. After 4 weeks, total lengths of elongated internodes were measured. In the absence of GA3, rice plants did not form elongated internodes after 4 weeks.
α-Amylase Induction in Embryoless Half Seeds of Rice.
Preparation of embryoless half seeds and induction of α-amylase were performed as described previously (9). The activity of α-amylase was assayed as described by Yamaguchi et al. (10). For the agar plate assay of α-amylase induction, 24 embryoless half seeds per plate were sterilized, washed, and positioned perpendicularly on a starch plate (0.2% starch and 2% agar) without GA3. The plates were incubated in the dark for 4 days at 30°C and then placed in a box saturated with iodine vapor. After a few minutes, the reaction between starch and iodine turned the agar plates a blue-purple color. The agar around half seeds with α-amylase activity remained colorless because of hydrolysis of starch by α-amylase.
For RNA gel blot analysis, 50 embryoless half seeds were placed in one well of a six-well titer plate. The seeds were sterilized, washed, and incubated in 2 ml of culture medium supplemented with 10−7 M GA3 for the indicated period at 30°C. After incubation, the embryoless half seeds were collected and stored at −80°C until RNA extraction.
Sequence Analysis.
Sequence analysis of the defective D1 gene in the d1 mutant was performed as described previously (4). The numbering of nucleotides corresponds to that submitted to GenBank (accession no. D38232).
RNA Gel Blot Analysis.
For RNA blot analysis, four GA-regulated genes were used as probes. Three were GA-inducible genes: Ramy1A, GAMYB, and Ca2 +-ATPase. Ramy1A (kindly provided by J. Yamaguchi, Nagoya University) encodes the most abundant GA-inducible α-amylase in the embryo. The DNA fragment corresponded to the GA-inducible MYB cDNA (OsGAMYB) and was prepared by a reverse transcription–PCR technique (11) with total RNA from young seedlings of rice. The DNA fragment contained the sequence from 1,008 to 1,743, which was confirmed to be identical to the reported sequence (12). The expression of a clone encoding a putative Ca2+-ATPase (expressed sequence tag clone, GenBank accession no. C62588) was induced in the aleurone layer by GA3 treatment as described by Wu et al. (13). The deduced amino acid sequence of the clone showed 82% identity to Wu's clone. The clone also displayed high similarity to the tomato endoplasmic reticulum-type (ER) ATPase (83% identity at the amino acid level) and to animal sarcoplasmic/endoplasmic reticulum Ca2+-ATPase (69%) within the B + C regions, which include five of the eight putative transmembrane domains essential for Ca2+ binding (14, 15). It was less similar to tomato plasma membrane-type H+-ATPase (21% identity) (16). On the basis of these findings, we concluded that the clone is a member of the GA-inducible ER-type Ca2+-ATPase gene family and used it as a cDNA probe for analyzing GA signaling in the aleurone layer. The cDNA encoding GA C20 oxidase (Os20ox), which catalyzes active GA biosynthesis, was also used as a probe [a kind gift from Y. Kamiya, RIKEN (17)].
These four cDNA probes were labeled with α32P-dCTP by the random priming method according to the manufacturer's instructions (Amersham). Extraction of total RNA from embryoless half seeds and internodes was performed as described previously (18, 19). Total RNA samples (10 μg for detection of OsGAMYB, Ca2+-ATPase and OsC20ox expression and 2 μg for detection of Ramy1A expression) were run on an agarose gel and transferred to Hybond N+ membrane (Amersham). Hybridization was performed at 65°C in ×5 standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), ×5 Denhardt's solution/0.5% SDS/10% dextran sulfate/0.1 mg/ml denatured salmon sperm DNA. Filters were washed with ×2 SSC/0.1% SDS at room temperature and then further washed twice with ×2 SSC/0.1% SDS at 65°C for 30 min and once with ×0.2 SSC/0.1% SDS at 65°C for 10 min.
Analyses of GA Content.
The quantitative analyses of endogenous GAs were performed by gas chromatography–selected ion monitoring with the procedure described previously (20).
Results
Phenotype of d1.
A d1 line, T65d1, was used to analyze the response to GA in the d1 mutant. In T65d1, a 2-bp deletion occurs at the positions of 1,003 and 1,004 in the D1 ORF, which results in frameshift mutation. The T65d1 mutant has a stop codon before the third effector-binding region (7) and therefore lacks this whole receptor-binding region. Thus, this allele should be a null mutant.
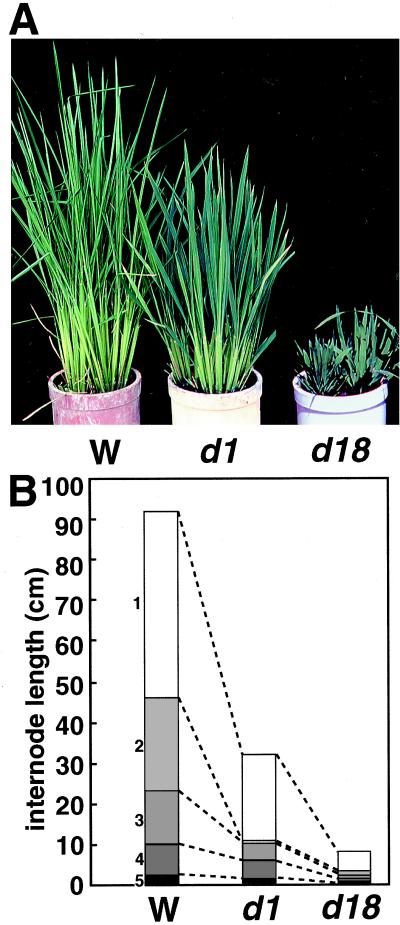
Fig. 1A shows the vegetative shoots of T65d1, compared with the wild-type and a GA-deficient mutant, d18, which is a loss-of-function mutant of GA 3β-hydroxylase in the GA synthetic pathway (H. Itoh, M.U.-T., N. Sentoku, H.K., M.M., and M.K., unpublished data). Although the d1 mutant was not as short as the d18 mutant, these mutants showed some similar phenotypes, namely wider leaf blades and darker green leaf sheaths and blades than those of the wild-type plants. These phenotypes are common to other mutants deficient in GA biosynthesis.
Figure 1.
(A) Phenotypes of d1 (Center), wild-type (Left), and d18 (Right) plants at 6 weeks. (B) Comparison of internode lengths of d1 (Center), wild-type (Left), and d18 (Right). Data are average internode lengths of 10 plants.
Fig. 1B shows a comparison of the internode elongation patterns of d1 with that of the wild-type and d18 plants. All internodes of d1 and d18 were shorter than those of the wild type, but the first (uppermost) internodes in both mutants were less affected than the other internodes. When the lower internodes of d1 and d18 were compared, the second internode was preferentially shortened in d1.
Inhibition of GA-Inducible Genes in Aleurone Layer of d1.
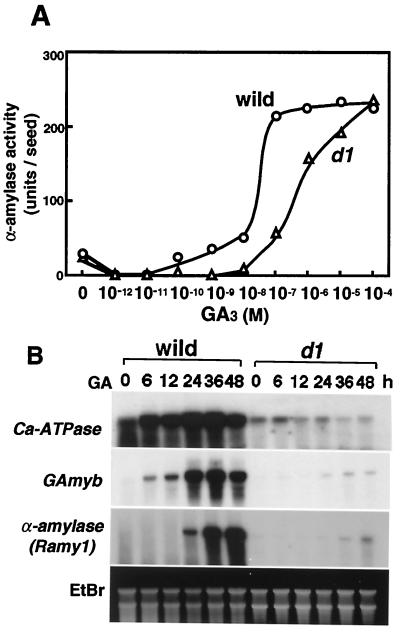
To investigate the possibility that the heterotrimeric G protein participates in the GA-signaling pathway leading to α-amylase expression, the induction of α-amylase activity in the d1 aleurone layer was analyzed (Fig. 2A). In the wild-type plant, α-amylase activity was induced at a concentration of 10−10 M GA3, and its induction was almost saturated at 10−7 M GA3. In the d1 mutant, however, 10−8 M GA3 did not induce enzyme activity. α-Amylase activity was gradually induced from 10−7 M to 10−4 M and finally reached a level similar to that of the wild type at around 10−4 M, which is much higher than the physiological GA level (5 ≈ 10 pg/seed ≒10−9 ≈ 10−8 M) in vivo (21).
Figure 2.
(A) GA induction of α-amylase activity in d1 and wild type. Embryoless half seeds were incubated for 4 days at 30°C in culture medium containing various concentrations of GA3 and then assayed for α-amylase activity as described in Materials and Methods. (B) RNA gel blot analyses of GA-inducible genes in aleurone cells. Fifty embryoless half seeds were incubated at 30°C for 0, 6, 12, 24, 36, and 48 h in culture medium containing 10−7 M GA3. Total RNA was extracted and analyzed by RNA gel blotting. Ten micrograms of total RNA for detection of GAMYB and Ca2 +-ATPase and 2 μg for detection of Ramy1 was loaded onto each lane. The gel was stained with ethidium bromide (EtBr).
This dose-response experiment showed that GA3 induction of α-amylase activity in the aleurone layer was more severely defected in the d1 mutant than in the wild-type plant. Given that the endogenous active GA level is around 10−9 M in embryos, the Gα-mediated pathway should be essential for α-amylase induction under natural conditions. However, the d1 mutant was still able to induce α-amylase activity when exposed to a high concentration of GA3 (about 10−7 M or higher), suggesting the presence of a GA-signaling pathway that is independent of the Gα protein (see Discussion).
To further investigate the GA-signaling pathway in the d1 aleurone layer, the expression of GA-inducible genes was analyzed. Ramy1A encodes the GA-induced α-amylase. The OsGAMYB gene is an ortholog of the barley GA MYB gene, whose product is thought to directly interact with the promoter of the GA-inducible α-amylase gene and positively regulate its expression (22). Rice Ca2 +-ATPase is homologous to the gene encoding the endoplasmic reticulum-vesicular localized Ca2+-ATPase (15), which is considered to be associated with the Ca2+ localization that is induced by GA signaling. Wu et al. (13) have reported that this type of ATPase is induced by GA treatment more rapidly than α-amylase. As shown in Fig. 2B, an increase in the transcripts of Ca2 +-ATPase and OsGAMYB was observed in the wild-type aleurone layer in the first 6 h after GA3 treatment and reached a maximum after 24 h, and the induction of α-amylase started at 24 h after treatment and reached a maximum after 36 h. Conversely, in the d1 aleurone layer treated with the same concentration of GA3, OsGAMYB and Ramy1 expression increased very slightly, and the response was much less than that observed in the wild type. There was no increase in expression of Ca2+-ATPase in d1. These RNA gel blot analyses indicate that GA signal transduction in the d1 aleurone layer is severely disrupted in terms of the induction of the GA-inducible genes.
Effect of GA on Shoot Elongation.
The role of the D1 product in GA signaling during shoot elongation was analyzed.
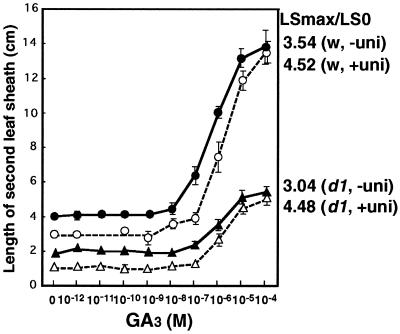
When the second leaf sheaths of d1 and wild-type plants were compared, the d1 seedlings were found to be less responsive to exogenous GA3 treatment than the wild type (Fig. 3). However, the elongation ratios were similar for d1 and wild-type plants at 3.04 and 3.54, respectively. To eliminate the effect of endogenous GA, uniconazol, inhibitor for GA biosynthesis, was used to pretreat the plants, and then the response to exogenous GA3 was investigated. Almost the same responsiveness for GA3 (LSmax/LS0 = 4.48 for d1 and 4.52 for wild-type plants) was observed. These results suggest that the Gα subunit may not play a large role in the second leaf sheath elongation in the vegetative seedlings. It may be noteworthy that the range of GA3 concentrations (10−8 to 10−4 M) used for leaf sheath elongation in both d1 and wild-type plants in this experiment was much higher than that normally used for the induction of α-amylase or other genes (10−10 M to 10−7 M).
Figure 3.
Elongation of the second leaf sheath in response to GA treatment in d1 (triangle) and the wild plant (circle) pretreated with (▵, ○) and without uniconazol (▴, ●). LSmax/LS0 is the ratio of the second leaf sheath length with GA3 treatment at 10−4 M (LSmax) to the length without GA3 treatment (LS0). n = 10.
Expression of GA biosynthetic enzymes such as GA C20 oxidase and GA 3β-hydroxylase is known to be suppressed by GA. It has been proposed that defects in the feedback suppression of the GA biosynthetic enzymes and accumulation of GAs are good criteria for determining whether the GA signal transduction pathway is functional (23). Thus, we analyzed the Os20ox expression in the second leaf sheath of the d1 mutant. RNA gel blot analysis revealed no great difference in the Os20ox expression level between d1 and wild plants (data not shown). Taken together, these results suggest that GA signaling at second leaf sheath may depend only slightly on the Gα protein.
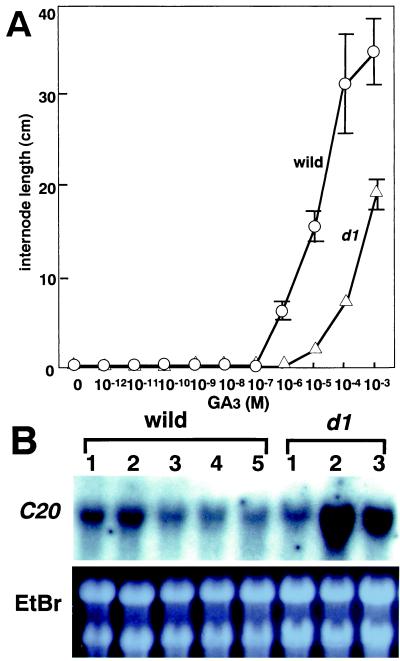
Under natural conditions in rice, internode elongation occurs only at the phase change of the shoot apical meristem from the vegetative to reproductive stage. However, exogenous addition of GA3 can induce internode elongation even in the juvenile phase. We used this phenomenon to assess the role of GA in internode elongation (Fig. 4A). In the wild type, internodes began to elongate at 10−7 M to 10−6 M GA3, and the response was almost saturated at a concentration of 10−4 M. In the d1 mutant, no internode elongation occurred at 10−6 M GA3 but there was a gradual increase in the internode length from 10−5 M to 10−3 M GA3. Thus, the effect of GA on internode elongation was about 100 times lower in the d1 mutant than in the wild type. It is noteworthy that higher concentrations of GA3 could induce internode elongation in the d1 mutant, as was the case for α-amylase induction.
Figure 4.
(A) Internode elongation in response to GA treatment in d1 and the wild type. (B) Expression of Os20ox in the first to third internodes of d1 and first to fifth internodes of wild-type plants. Total RNA was extracted from each internode and analyzed by RNA gel blotting. Ten micrograms of total RNA was loaded onto each lane, and the gel was stained with ethidium bromide (EtBr).
The response of internode elongation to exogenously applied GA3 was very different in the d1 mutant and wild-type plants. It is possible that this effect may be reflected under natural conditions by differences in the response to endogenous GA. The most severely affected internode in the d1 mutant was the second internode, whereas the elongation of the first internode was much less affected. The first to fifth internodes from the wild-type and the first to third internodes from d1 plants were individually collected at the stage when the first and second internodes were actively elongating. At this time, elongation of the fifth internode has finished, and elongation of the third and fourth internodes is almost complete. Total RNA was extracted from each internode of wild-type and d1 plants and tested for the Os20ox expression (Fig. 4B). As expected, moderate expression of the gene in the wild type was observed in the first and second internodes, which were actively elongating at sampling time, whereas lower expression was seen in the internodes that had already stopped elongation. In the d1 mutant, the expression level was increased almost 6 times higher and 4.5 times higher in the second and third stunted internodes, respectively, compared with the wild type, whereas its expression was almost similar or slightly lower in the first internode. The increased level of Os20ox expression in the severely stunted internodes indicates that the feedback suppression of Os20ox by active GAs does not function in these shortened internodes.
The high-level expression of Os20ox leads us to the possibility that higher levels of active GAs are accumulated in these internodes. To test this possibility, we directly measured the amount of GAs in these internodes and compared those to the wild plants (Table 1). As we expected, each amount of GA20 and GA1 in the stunted internodes of d1 was five times higher and three times higher than that of the wild plant, respectively.
Table 1.
Levels of endogenous GAs in the internodes of wild-type and d1 mutants, nanograms/grams fresh weight
| GA53 | GA44 | GA19 | GA20 | GA1 | GA8 | GA29 | ||
|---|---|---|---|---|---|---|---|---|
| W | 2nd | 0.96 | 0.63 | 4.80 | 0.91 | 0.27 | 0.26 | 0.39 |
| 3rd | 0.64 | 0.23 | 1.00 | 0.97 | 0.45 | ND | 0.46 | |
| d1 | 2nd | 3.36 | 2.90 | 8.70 | 4.57 | 0.76 | ND | 2.21 |
| 3rd | 0.74 | 0.62 | 5.40 | 2.03 | 2.03 | ND | 0.94 |
Epistatic Analysis of the d1 and slender rice Mutants.
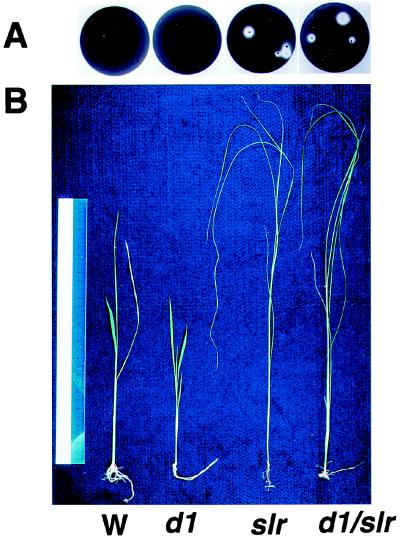
In rice, there is another GA-related recessive mutant, slender rice (slr), which shows GA constitutive response phenotypes including the slender phenotype. Recently, we found that the slr mutant contains a defect in the function of a rice GAI (24) or RGA (25) homologous gene, which plays as a negative regulator in GA signaling in Arabidopsis (Ikeda et al., personal communication). To determine the epistatic relationship between d1 and slr, the two mutants were crossed. Because the homozygous plant of slr is sterile, the heterozygous plant of slr was crossed with a d1 homozygous plant. The genotype of F2 plants was directly identified by sequence analysis of D1 and SLR. The α-amylase activity of the mutants was examined by using the starch-plate assay system (Fig. 5A). Embryoless half seeds of the homozygous slr mutant displayed α-amylase activity without addition of GA3, whereas those of the wild type and d1 showed no α-amylase activity in the absence of GA3. Embryoless half seeds of the double mutant (slr/d1) gave the same result as the slr single mutant. Fig. 5B shows the overall phenotypes of seedlings of the wild type, single d1, single slr, and double mutant of slr/d1 at 2 weeks. The double mutant showed a “slender” phenotype, that is, pale green and elongated leaf sheaths and leaf blades. Furthermore, the double mutants grown for 50 days showed internode elongation the same as the slr single mutant, whereas both wild and d1 plants at the same stage never elongated internodes (data were not shown). These results demonstrate that slr is epistatic to d1.
Figure 5.
Epistatic analysis of d1 and slr mutants. (A) A plate assay of α-amylase induction. Embryoless half seeds of the wild type (Left), d1, slr, and d1/slr (right) were sterilized, washed, and incubated on starch plates without GA3 for 4 days at 30°C. (B) Two-week-old seedlings of the wild type (W), d1, slr, and d1/slr.
Discussion
The GA-signaling pathway is partially impaired in the rice dwarf mutant, d1, which is defective in the Gα protein. The defect in the GA-signaling pathway in d1 is evident in organ and/or GA responsive events, in particular, the induction of α-amylase activity and expression of GA-inducible genes in aleurone. Internode elongation is also severely affected in the d1 mutant. Physiological experiments with a constitutive activator of the heterotrimeric G protein, Mas7, indicate that the Gα protein may be involved in the GA-signaling pathway in higher plants (6). However, because Mas7 also activates other signal compounds such as calmodulin, phospholipase C, and small G proteins (26), further evidence for the direct involvement of the Gα protein in GA signaling is required. In this study, we clearly demonstrate that the Gα protein is essential for the induction of α-amylase and other GA-inducible genes in aleurone under normal physiological conditions. Furthermore, we have shown that the phenotypes of d1 are completely suppressed in a d1/slr double mutant, which instead shows phenotypes identical to that of the slr mutant. The slr mutant is caused by the loss of function of a rice GAI or RGA homologue, a negative regulator of GA signaling (Ikeda et al., personal communication; refs. 24 and 25). Consequently, the identical phenotypes in the single slr and double slr/d1 mutants indicate that the D1 product functions as a member of the same GA-signaling pathway as the SLR protein.
Although d1 plants are severely affected in the GA-signaling pathway for aleurone and internode elongation, the plants do not completely lose all GA signaling. Indeed, d1 shows a GA-responsive reaction of the second leaf sheath elongation similar to that of wild-type plants. In addition, α-amylase activity and internode elongation in d1 are induced by high levels of GA3. We do not believe that these GA-responsive reactions in d1 are because of the functional redundancy of the rice Gα subunit or partial functioning of the D1 protein in d1. DNA gel blot analysis of the rice genome has shown that even under the low-stringency conditions, there is only one DNA fragment that hybridizes with the Gα cDNA clone (4). Moreover, Arabidopsis may contain only one gene encoding the Gα subunit, because we have found only one gene in a blast search of the Arabidopsis genome. It is also unlikely that the D1 product partially maintains its function in the d1 mutant, because the D1 gene in the mutant produces a truncated product that lacks some domains essential for its function. Furthermore, four other d1 mutants with mutations at different positions have the same abnormal phenotype (data not shown). On the basis of these observations, we believe that the Gα protein is the single gene in rice and that the d1 mutant used in this study is a nonfunctional mutant of D1 that encodes the Gα protein.
Why then does d1 show only partial impairment of GA signaling? As described above, α-amylase induction in the aleurone layer is severely affected in d1 at low concentrations of GA3. It is possible that Gα may commit to GA signaling at low concentrations of GA3 (10−10 M to 10−7 M). However, α-amylase activity at a level similar to that in the wild-type plant is observed in d1 at around 10−4 M GA3, which is a much higher concentration than physiological GA levels in vivo. This suggests that there may be an alternative pathway(s) for high concentrations of GA3 that does not involve Gα (Fig. 6A). Second leaf sheath elongation in vegetative seedlings of the d1 mutant is not largely affected, but the concentration of GA (10−8 M to 10−4 M) required for leaf sheath elongation is higher than that needed for α-amylase induction (10−10 M to 10−7 M). This also indicates the presence of an alternative signaling pathway for response to high concentrations of GA without Gα commitment. Nakajima et al. (27) have provided evidence for the occurrence of a cytosolic GA-binding protein in the GA-responsive elongation in Azuki seedling. It is possible that an alternative GA-signaling pathway might involve such a cytosolic receptor. GA stimulation of internode elongation in d1 is also affected, and the elongation in d1 internode occurs at a higher concentration of GA3 compared with that required for α-amylase induction. This may be caused by the artificial assay system by using vegetative seedlings, because no internode elongation occurs in rice plants at the vegetative stage under normal growth conditions.
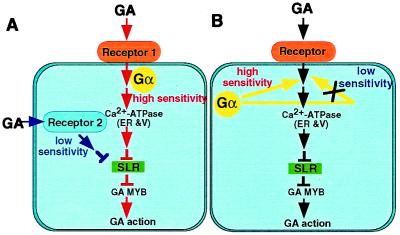
Figure 6.
Diagram summarizing the two proposed GA-signaling pathways in rice. (Model A) Gα works in high sensitive GA receptor system. There may be an alternative GA receptor system for low sensitivity to GA that does not involve Gα. High sensitive α-amylase induction and internode elongation by GA may be mediated mainly through the high sensitive reception system (red), whereas leaf sheath elongation may be through the low sensitive reception system (blue). (Model B) Gα works in a pathway that regulates the GA signaling. α-Amylase induction and internode elongation may be regulated mainly by this pathway to result in high sensitivities to GA (red), whereas leaf sheath elongation may be not regulated by this pathway to result in low sensitivity to GA (blue). The SLR protein works at the downstream site of these two pathways.
Multiple signaling pathways are known for perception of other phytohormones. For example, it has been reported that multiple abscisic acid (ABA) signaling pathways may be involved in the different ABA dose–response curves observed for Em∷GUS and C1∷GUS in maize protoplasts (28). The dose-response curve for ABA activation of GUS activity under the control of the Em promoter in maize protoplasts is nonsaturable over a broad range of ABA concentrations (10−7 M to 10−3 M). In contrast, ABA activation of GUS activity controlled by the C1 promoter reaches a maximum at 10−5 M ABA, which is consistent with the fact that C1 is coupled to a single high-affinity saturable system. Multiple perception systems linked to parallel signal transduction pathways could provide a strategy for modulating the sensitivity of different physiological processes in cells to the same initial stimulus.
Another interpretation for partial impairment of GA-signaling pathway in d1 mutant is that the Gα protein may act in a pathway that regulates the GA signaling (Fig. 6B). GA-dependent α-amylase induction and internode elongation may be regulated mainly by this pathway to result in high sensitivities to GA, whereas the second leaf sheath elongation may be not regulated to result in low sensitivity to GA.
Although the second leaf sheath elongation in response to GA is almost the same in d1 as in the wild type, d1 plants have shorter leaf sheaths than the wild type. At present, we do not have a clear answer to explain the shorter leaf sheath of d1. However, defects in Gα-mediated GA signaling may affect the cell numbers in this organ. In this case, the Gα-mediated GA signaling would regulate division of leaf sheath cells but not elongation. This interpretation is consistent with the previous finding that the cell numbers in d1 seedlings are reduced compared with the wild type (29).
Acknowledgments
This work was supported by the “Research for the Future” Program from The Japan Society for the Promotion of Science (FSPS-RFTF 96L00601) and by a grant-in-aid for Scientific Research on Priority Areas (The Molecular Bases of Flexible Organ Plans in Plants) from the Ministry of Education, Science and Culture (Japan).
Abbreviations
- GA
gibberellin
- ABA
abscisic acid
- Gα
α subunit of the heterotrimeric G protein.
References
- 1.Ma H, Yanofsky M F, Meyerowitz E M. Proc Natl Acad Sci USA. 1990;87:3821–3825. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma H, Yanofsky M F, Meyerowitz E M. Gene. 1991;107:189–195. doi: 10.1016/0378-1119(91)90318-6. [DOI] [PubMed] [Google Scholar]
- 3.Poulsen C, Mai X M, Borg S. Plant Physiol. 1994;105:1453–1454. doi: 10.1104/pp.105.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa A, Tsubouchi H, Iwasaki Y, Asahi T. Plant Cell Physiol. 1995;36:353–359. doi: 10.1093/oxfordjournals.pcp.a078767. [DOI] [PubMed] [Google Scholar]
- 5.Gotor C, Lam E, Cejudo F J, Romero L C. Plant Mol Biol. 1996;32:1227–1234. doi: 10.1007/BF00041411. [DOI] [PubMed] [Google Scholar]
- 6.Jones H D, Smith S F, Desikan R, P-, Dymock S, Lovegrove A, Hooley R. Plant Cell. 1998;10:245–253. doi: 10.1105/tpc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y. Proc Natl Acad Sci USA. 1999;96:7575–7580. doi: 10.1073/pnas.96.13.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Proc Natl Acad Sci USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi J. Breeding Sci. 1998;48:365–370. [Google Scholar]
- 10.Yamaguchi J, Itoh S, Saitoh T, Ikeda A, Tashiro T, Nagato Y. Theor Appl Genet. 1999;98:32–38. [Google Scholar]
- 11.Huang N, Koizumi N, Reinl S, Rodriguez R L. Nucleic Acids Res. 1990;18:7007–7014. doi: 10.1093/nar/18.23.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubler F, Watts R J, Kalla R, Matthews P, Keys M, Jacobsen J V. Plant Cell Physiol. 1997;38:362–365. doi: 10.1093/oxfordjournals.pcp.a029175. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Chang M, Wang B, Wu R. Plant J. 1997;11:363–371. doi: 10.1046/j.1365-313x.1997.11030363.x. [DOI] [PubMed] [Google Scholar]
- 14.Andersen J P, Vilsen B. Curr Opin Cell Biol. 1990;2:722–730. doi: 10.1016/0955-0674(90)90116-v. [DOI] [PubMed] [Google Scholar]
- 15.Wimmers L E, Ewing N N, Bennet A B. Proc Natl Acad Sci USA. 1992;89:9205–9209. doi: 10.1073/pnas.89.19.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing N N, Wimmers L E, Meyer D J, Bennett A B. Plant Physiol. 1990;94:1874–1881. doi: 10.1104/pp.94.4.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyomasu T, Kawaide H, Sekimoto H, Von Numers C, Phillips A L, Hedden P, Kamiya Y. Physiol Plant. 1997;99:111–118. [Google Scholar]
- 18.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 19.Perata P, Geshi N, Yamaguchi J, Akazawa T. Planta. 1993;191:402–408. [Google Scholar]
- 20.Kobayashi M, Gomi M, Agematsu J, Asami T, Yoshida S, Sakurai A. Biosci Biotechnol Biochem. 1995;59:1969–1970. [Google Scholar]
- 21.Choi Y-H, Kobayashi M, Sakurai A. J Plant Growth Regul. 1996;15:147–151. [Google Scholar]
- 22.Gubler F, Kalla R, Roberts J K, Jacobsen J V. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornton T M, Swain S M, Olszewski N E. Trends Plant Sci. 1999;4:424–428. doi: 10.1016/s1360-1385(99)01485-5. [DOI] [PubMed] [Google Scholar]
- 24.Peng J, Carol P, Richards D E, King K E, Cowling R J, Murphy G P, Harber d N P. Gene Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverstone A L, Ciampaglio C N, Sun T P. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho M H, Tan Z, Erneux C, Shears S B, Boss W F. Plant Physiol. 1995;107:845–856. doi: 10.1104/pp.107.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima M, Takita K, Wada H, Mihara K, Hasegawa M, Yamaguchi I, Murofushi N. Biochem Biophys Res Commun. 1997;241:782–786. doi: 10.1006/bbrc.1997.7896. [DOI] [PubMed] [Google Scholar]
- 28.McCarty D R. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- 29.Kamijima O. Jpn J Breed. 1981;31:302–315. [Google Scholar]