Abstract
Disease resistance is associated with a plant defense response that involves an integrated set of signal transduction pathways. Changes in the expression patterns of 2,375 selected genes were examined simultaneously by cDNA microarray analysis in Arabidopsis thaliana after inoculation with an incompatible fungal pathogen Alternaria brassicicola or treatment with the defense-related signaling molecules salicylic acid (SA), methyl jasmonate (MJ), or ethylene. Substantial changes (up- and down-regulation) in the steady-state abundance of 705 mRNAs were observed in response to one or more of the treatments, including known and putative defense-related genes and 106 genes with no previously described function or homology. In leaf tissue inoculated with A. brassicicola, the abundance of 168 mRNAs was increased more than 2.5-fold, whereas that of 39 mRNAs was reduced. Similarly, the abundance of 192, 221, and 55 mRNAs was highly (>2.5-fold) increased after treatment with SA, MJ, and ethylene, respectively. Data analysis revealed a surprising level of coordinated defense responses, including 169 mRNAs regulated by multiple treatments/defense pathways. The largest number of genes coinduced (one of four induced genes) and corepressed was found after treatments with SA and MJ. In addition, 50% of the genes induced by ethylene treatment were also induced by MJ treatment. These results indicated the existence of a substantial network of regulatory interactions and coordination occurring during plant defense among the different defense signaling pathways, notably between the salicylate and jasmonate pathways that were previously thought to act in an antagonistic fashion.
Active disease resistance in plants depends on the ability of the host to recognize pathogens and initiate defense mechanisms that limit infection. Resistance in the host is often manifested by a hypersensitive response, which results in localized cell death at the site of infection. Other defense responses may include structural alterations and the production of a wide range of plant defense molecules such as antimicrobial proteins (see refs. 1–3 for recent reviews). In addition, plant responses to necrotrophic pathogens can lead to systemic acquired resistance, which immunizes against subsequent infection. Endogenous signal molecules such as salicylic acid (SA) play a key role in signaling for resistance (4, 5). Recently, signal transduction pathways that involve jasmonates and ethylene as regulators of several defense-related genes have also been identified (6–11). Crosstalk between defense pathways mediated by salicylates, jasmonates, ethylene, and pathogen infection has been proposed (7, 9). However, the analysis of signaling processes and their interactions in plants have traditionally focused on only one or a few genes at any one time (12). From such studies it has not been possible to assess the extent of overlap of gene activation by different signals and pathogens in the defense response. Quantitative methods for global and simultaneous analysis of expression profiles, such as the recently developed cDNA microarray analysis, can improve our overall understanding of the molecular basis of the plant defense response. The measurement of expression levels of thousands of genes in parallel serves as an important tool in functional genomics, and the expression profiles of genes with no known function obtained by microarray analyses have been useful for assigning putative roles (13–16). In the present study, we examined the changes that occur in the abundance of transcripts corresponding to 2,375 Arabidopsis expressed sequence tags (ESTs) with a biased representation of putative defense-associated and regulatory genes after inoculation with an incompatible pathogen or treatment with low molecular weight signal compounds. Our results demonstrated the existence of a substantial network of regulatory interaction and coordination occurring among different plant defense pathways.
Materials and Methods
Treatments of Arabidopsis Plants.
Arabidopsis thaliana Columbia plants (8–12 leaf stage) grown in autoclaved potting mix in controlled environment rooms at 24–20°C day and night temperature and a photoperiod of 16 h light (500 μE/m2/sec) were used for each treatment and experimental replicate. Alternaria brassicicola (isolate UQ4273) was grown on agar plates containing clarified V8 vegetable juice (Campbell Soup Co., Camden, NJ). For inoculations, 5-μl drops of a spore suspension (5 × 105 spores/ml in water) were pipetted onto two to four leaves per plant (one to two drops per leaf). The plants were then placed in a 20-liter container with a clear polystyrene lid and kept at high humidity. Control plants were not inoculated, but were otherwise treated in the same way. For treatments with SA, plants were sprayed with a 4-mM solution. Methyl jasmonate (MJ) treatments were carried out by taping a cotton ball containing 400 μl of a 0.5% solution in ethanol onto the wall of a 20-liter container with a clear polystyrene top wrapped in plastic bags. For treatments with ethylene, 10 ml was injected into the air of plants kept under a sealed 50-liter Plexiglas box on a laboratory bench. The control plants were treated in the same way as the individual chemical treatments but without the addition of SA, MJ, or ethylene. Leaves of treated and untreated control plants were collected 24 h after chemical treatments and 72 h after Alternaria inoculation (leaves with visible traces of fungal spore inoculations). To confirm that A. brassicicola inoculation had effectively triggered plant defense responses, RNA samples isolated from untreated and Alternaria-inoculated leaves were first subjected to Northern blot analysis by using a cDNA (GenBank accession no. T04323) probe for the PDF1.2 gene of Arabidopsis. This gene encodes a plant defensin induced by fungal challenge (6). A strong band was observed for the RNA isolated from inoculated plants but not for the uninoculated leaves (data not shown).
Isolation of Total RNA and Northern Blot Analyses.
Total RNA for microarray or Northern blot hybridizations was isolated from 2–4 g Arabidopsis leaf material according to Chirgwin et al. (17). RNA blots were prepared by gel electrophoresis and blotting of total RNA as described by Sambrook et al. (18) by using Hybond N+ (Amersham) and ×10 SSC as the transfer buffer. cDNA probes were amplified by PCR with corresponding primers of the EST and labeled by using a Megaprime radiolabeling kit (Amersham). Hybridization and stringency washes were carried out according to manufacturers' instructions with the initial wash buffer being replaced with ×5 SSPE (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA).
Microarray Preparation.
Arabidopsis cDNA plasmid clones of 2,375 ESTs generated from the collections of Höfte et al. (19), Cooke et al. (20), and Newman et al. (21), and various other miscellaneous clones were collected. For control purposes, 148 ESTs were represented more than once on the microarray. The identity of 22% of the arrayed clones was confirmed by sequencing. Inserts were amplified by PCR under conditions outlined by Ruan et al. (22). PCR products were ethanol precipitated and resuspended in 10 μl of ×3 SSC buffer, and samples of each were visualized on 1% agarose gels to ensure PCR-amplification quality and quantity. The PCR fragments were arrayed from 384-well microtiter plates onto silylated microscope slides by CEL Associates (Houston, TX) by using an Omnigrid Microarrayer from Genemachines (San Carlos, CA) with ChipMaker 2 pins from TeleChem International (Sunnyvale, CA). Postprinting slide procedures were performed as described in Heller et al. (23).
Fluorescent Probe Preparation, Hybridization, and Scanning.
mRNA was isolated from 100 μg total RNA (Qiagen Midi Kit, Chatsworth, CA) and reverse transcribed by using Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) and an oligo(dT) 23 mer, as recommended by the manufacturer. The resulting cDNA was treated with 1 unit of RNase H for 30 min at 37°C, purified by using a Centricon-30 filtration spin column (Amicon, Beverly, MA), and concentrated to <20 μl. One-tenth of the cDNA sample was then labeled with either Cy-3 or Cy-5-labeled dUTP (Amersham) by a randomly primed polymerization reaction. In brief, 20-μl labeling reactions contained cDNA, 2 μl of ×10 Klenow buffer (United States Biochemical), 0.5 μl of fluorescent dUTP (25 nmol), 3 μg of random primers (Life Technologies), 2 μl each of 250 μM dATP, dCTP, dGTP, 90 μM of dTTP, and 1 unit of Klenow enzyme (United States Biochemical). After incubation at 37°C for 3 h, the reactions of two samples (one with Cy-3 and one Cy-5) were combined and purified by using a Centricon-30 filtration spin column. The sample was then lyophilized and dissolved in 14 μl of hybridization buffer (×3 SSC/0.3% SDS/2.4 μg yeast tRNA/1.4 μg of sheared salmon sperm DNA). The probe was denatured at 99°C for 2 min, applied to the microarray, and covered with a 22 × 22 mm2 Hybrislip (Research Products International). The slide was then placed in a waterproof hybridization chamber for hybridization in a 65°C water bath for 12–16 h. After hybridization, slides were washed 2 min in each of ×1 SSC with 0.03% SDS/×0.2 SSC/×0.05 SSC. Slides were scanned with a ScanArray 3000 (GSI Lumonics, Oxnard, CA). Careful handling and prescanning of slide surfaces before hybridizations as well as thorough washing steps ensured a minimum of dust and salt precipitates.
Data Analysis.
Stringent control measures were applied for all steps of the data analysis so that all results on gene induction and repression presented were reproduced and had signals that were within the window of resolution of the microarray hybridization method. Control measures included quality controls of the microarray cDNA by gel analysis, the setup of at least two independent replicate experiments with corresponding untreated controls for each treatment, an additional background cutoff to eliminate genes with very low signals, and an induction or repression ratio cutoff of at least 2.50. A negative control hybridization by using two independent untreated control samples from the Alternaria inoculation experiment was used to estimate the expected level of experimental variation in measuring gene expression (Table 1). In this control experiment, 2.1% of all ESTs showed variation in mRNA levels >2.5-fold between the two independent controls. Assuming that this experimental variation is nonspecific to any particular EST, the use of two independent experimental replicates for data acceptance would reduce experimental variation and misassignment of gene induction to <0.05%. In case the ESTs that showed variation in transcript abundance between these controls represented transcripts especially sensitive to environmental parameters and that could not be adequately controlled, the data for these ESTs (by using cutoff ratios >2.00) was removed from the analysis of chemical treatments and infection. Because of the stringent criteria used for data acceptance, it is likely that some genes differentially expressed during the treatments may have been eliminated.
Table 1.
Overview of experiments and differential hybridizations used for microarray analysis
| Treatment | A. brassicicola | Salicylic acid | Methyl jasmonate | Ethylene | Negative control |
|---|---|---|---|---|---|
| Replicate hybridization comparisons | A1−/A1+ | SA1−/SA1+ | MJ1−/MJ1+ | Eth1−/Eth1+ | A1−/A2− |
| A2−/A2+ | SA2−/SA2+ | MJ2−/MJ2+ | Eth2−/Eth2+ | ||
| Extra experimental replicate | A2−/A3+ |
Different experimental replicates are shown as different numbers. Untreated/noninoculated controls are indicated by a minus (−) and treated/inoculated samples by a plus (+) sign. Treatments are as follows: A = Alternaria, SA = salicylic acid, MJ = methyl jasmonate, and Eth = ethylene.
For the data analysis, spot intensities from scanned slides were quantified by using imagene 2.0 software (Biodiscovery). Grids were predefined and manually adjusted to ensure optimal spot recognition, discarding spots with dust or locally high background. Spots were individually quantified by using imagene's fixed circle method; sample value was measured as the mean of pixels within a circle encompassing the spot and the local background in a four-pixel-wide torus that began two pixels outside the fixed circle. Gene expression data were normalized by using a set of custom Perl scripts. Further information on these scripts can be obtained from http://cellwall.stanford.edu/scripts/index.shtml and supplemental data at www.pnas.org. Briefly, data were normalized between channels and replicate slides by using the following procedure: normalized value S = χ/σ, where χ = sample value and σ = channel standard deviation (calculated ignoring the upper 2.5% and lower 2.5% of the data). Overall background for each experiment was calculated by using a set of 118 control spots on each slide. Data points where expression was not greater than two standard deviations above the overall background for at least one channel were discarded. Anomalous data points missed by manual inspection were removed automatically when replicate spots varied by more than 10-fold in value. Furthermore, genes that did not show a single strong PCR band on agarose gels before slide printing were also not considered for further analyses. It is expected that weak hybridization signals are less reproducibly quantified than stronger signals. Therefore, an additional background criterion was applied: for the highest of the two signals, only signals that were higher than the average background (plus ×2 SD) of that channel and that were also at least 2-fold higher than the average background (plus ×2 SD) of the other channel (displaying the lower signal) were considered for further data analysis. For the final analysis, data points were averaged from two replicates for cluster analyses to provide equal parameters for comparative analyses (for details of Perl scripts used, see http://cellwall.stanford.edu/scripts/index.shtml). Data obtained from a third experimental replicate (Table 1) for the Alternaria treatment were incorporated into Fig. 3 to further increase the significance of data obtained for the Alternaria inoculation. Analyses of all combinations of induction or repression profiles of two experimental replicates were systematically carried out by using graphical approaches and the sort and count-if functions of Microsoft Excel. The ability of the microarray method to reproducibly detect chemical-induced changes in gene expression was tested by including multiple cDNAs on the array, for example PR1, a well-characterized SA-inducible marker gene. Three replicated spots for PR1 were included on the array (including one from a related Arabis species; CI0007), and induction was observed in plants treated with SA with mean induction ratios of 5.6, 7.8, and 11 recorded for the PR1 spots on the array in the two experiments.
Figure 3.
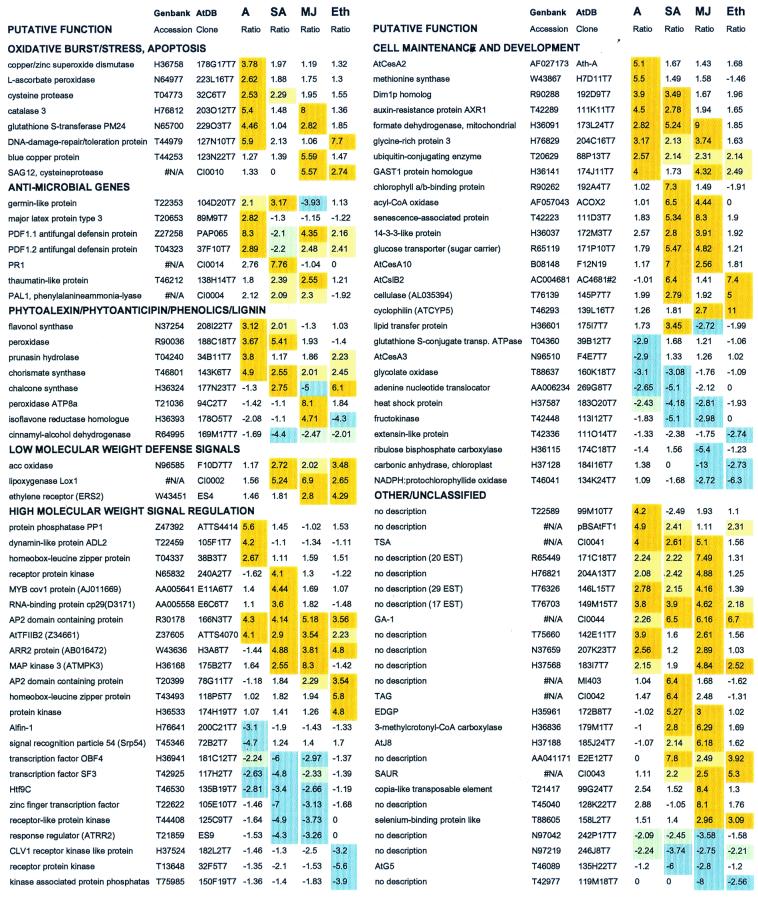
Examples of expression profiles of genes that are significantly induced (positive ratios, shaded in light orange) or repressed (negative ratios, shaded in light blue) by Alternaria (A), SA, MJ, or ethylene (Eth) treatments. Listed by functional groups are the 20 genes that were most highly induced by Alternaria inoculation and the 10 genes that were most highly induced by each of the signal chemical treatments, as well as the five most repressed genes for each treatment. In addition, some other selected genes that showed multiple induction or repression profiles are shown. Ratios between 2.00 and 2.50 that matched all other criteria for data analysis (except for the ratio cutoff) are shaded in light yellow for induced and light green for repressed genes. Data obtained from an additional replicate experiment for the Alternaria inoculation was incorporated. N/A, not accessible. The complete list can be accessed at http://www.tpp.uq.edu.au/microarray/responsivegenes.htm or from supplemental data at www.pnas.org.
Results and Discussion
Array-Based Hybridization Experiments and Analysis of Expression Profiles.
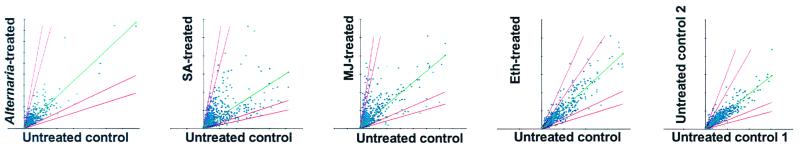
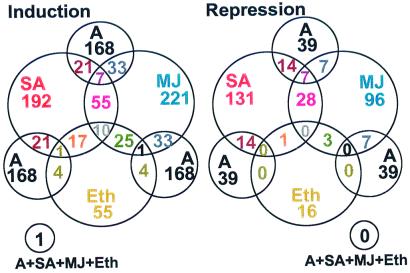
In this study, we examined the changes that occur in the abundance of transcripts corresponding to 2,375 Arabidopsis ESTs with a biased representation of putative defense-associated and regulatory genes. A. thaliana cv. Columbia plants were either inoculated with an incompatible strain of A. brassicicola or treated with the signal molecules SA, MJ, or ethylene. Total RNA was isolated from leaves of untreated controls and treated plants at 72 h after fungal inoculation or 24 h after chemical application. The experiments undertaken are described in Table 1. In this analysis, only changes in mRNA abundance in excess of 2.5-fold that of controls in all replicate experiments were accepted. Experimental replication, multiple controls, and stringent criteria for data analysis were used (see Materials and Methods). Analyses of these data revealed that 705 ESTs on the microarray showed significant differential expression in response to one or more of the treatments. The strongest overall response observed was for the SA and MJ treatments and the weakest, apart from the negative control, was for the ethylene treatment (Fig. 1). After fungal inoculation, transcript levels of 168 genes were increased whereas those of 39 genes were decreased (Fig. 2). Similarly, the transcript abundance of 192 genes for SA, 221 genes for MJ, and 55 genes for ethylene was increased as a result of treatment with these signal molecules. In contrast, transcript abundance of 131, 96, and 16 genes was reduced after treatment with SA, MJ, and ethylene, respectively (Fig. 2). Fig. 3 shows examples of genes with both high induction/repression ratios for individual treatments and high ratios across multiple treatments.
Figure 1.
Overview of experiments and scatter plot graphs of expression distribution patterns of 2375 ESTs after microarray hybridizations with labeled cDNA probes obtained from mRNA of untreated and treated Arabidopsis plants (average of two replicates for each treatment). Treatments with corresponding untreated controls included inoculation with A. brassicicola and applications of the defense-related signaling molecules SA, MJ, and ethylene (Eth). A negative control hybridization was carried out with two untreated control samples. Diagonal red lines represent 2-fold and 3-fold induction/repression ratio cutoffs relative to the best fit line through the normalized data (middle green line).
Figure 2.
Venn diagrams of the numbers of overlapping and nonoverlapping induced or repressed genes on the array with ratios of at least 2.50 after inoculation with A. brassicicola (A) or treatments with defense-related signal molecules [SA, MJ, or ethylene (Eth)] based on two experimental replicates. A complete list of genes from all 2,375 ESTs with ratios obtained from data analysis of two experimental replicates is accessible on the web site http://www.tpp.uq.edu.au/microarray/clusterdata.htm or from supplemental data at www.pnas.org.
Functional Classification of Genes with Altered Expression Patterns.
Approximately 10% of responsive genes had known functions or had previously been implicated in plant defense, whereas approximately 35% were involved in cell maintenance or development. The first four functional groups given in Fig. 3 included genes that have been reported to have defensive roles as well as genes implicated in the plant defense response. Transcript abundance of these genes, as expected, was mostly up-regulated by at least one of the treatments. The first group contained genes that are implicated in the oxidative burst and programmed cell death or hypersensitive response. The oxidative burst may be followed by activation of genes encoding antioxidant enzymes in the tissue surrounding the initial infection site (24), such as catalases and glutathione S-transferase (GST1) as well as a putative DNA repair protein (Fig. 3). A gene encoding a putative cysteine protease was induced by fungal inoculation and MJ and ethylene treatments. Recently, cysteine proteases have been implicated as mediators of pathogen-induced cell death in other plants (25, 26). The second group (antimicrobial genes) included some of the previously known defense genes, such as PR1 and PDF1.2 (6), a thaumatin-like protein (27), and PAL1 (28). The expression pattern observed for PDF1.1 was similar to that of PDF1.2. This contradicts expression data obtained by Penninckx et al. (6) and is probably because of cross hybridization between these two closely related genes. Differentiating between closely related sequences is one limitation of current cDNA microarray systems, and arrays containing 3′ and 5′ untranslated regions will be needed to differentiate gene family members (29).
In this study, emphasis was given to genes putatively involved in signal recognition and transduction, and at least 25% of all genes with significant altered expression belong to this group (Fig. 3). For instance, MJ and ethylene treatments were associated with enhanced transcript accumulation for several putative regulatory genes, such as two genes encoding AP2 domain-containing proteins. The AP2-domain family of transcription factors interacts with ethylene responsive elements present in the promoters of ethylene inducible genes (30). Recently, MJ inducibility was also reported for an AP2-domain transcription factor (31). In this group, another gene, which was highly induced by MJ but to a lesser extent by SA, encodes a putative lipoxygenase (Lox1). Previously, induction of this gene by pathogen and MJ has also been reported (32). These results are consistent with coregulation of genes required for MJ synthesis and signaling. In addition, fungal inoculation induced the expression of genes encoding a protein phosphatase and a leucine zipper protein while repressing the expression of genes encoding a zinc-finger nucleic acid-binding protein and a GTP-binding protein. Ethylene treatment induced the expression of genes for a leucine zipper protein and a protein kinase, whereas it repressed the expression of genes for a CLV1 receptor kinase-like protein, a receptor protein kinase, and a kinase-associated protein phosphatase. SA and MJ treatments up-regulated the expression of genes encoding an AP2 domain-containing protein (T20399), an AtTFllB2 transcription factor, and a mitogen-activated protein kinase. In contrast, both treatments down-regulated the expression of genes for a zinc-finger transcription factor, a receptor-like protein kinase, and a response-regulator protein. Interestingly, both fungal inoculation and ethylene treatment down-regulated the zinc-finger proteins encoded by the two separate genes. Although assignment of a defensive function to these genes still awaits further experimentation, in light of recent evidence, involvement of genes encoding transcription factors with zinc-finger/binding domain in regulating plant defense responses appears to be very likely. The zinc-finger protein encoded by the Arabidopsis LSD1 gene that acts as a negative regulator of hypersensitive response to restrict the spreading of cell death is one of the examples of such a gene (33). These putative regulatory genes would potentially function in transmitting pathogen signals (34, 35) and thus could be useful as candidates for further functional studies. Such studies may include more detailed characterization of expression patterns (i.e., time-course studies) or over- or under- (or disruption) expression of the genes in transgenic plants.
At least 7% of responsive genes encode proteins associated with cell-wall synthesis and modification. The majority (approximately 80%) of these genes were induced rather than repressed on infection or signal chemical treatment. These induced genes include homologues of defense-related lignin biosynthetic enzymes such as peroxidase and cinnamyl alcohol dehydrogenase and structural glycine-rich and extensin proteins. The expression of cellulose synthases (e.g., AtCesA2, CesA3, AtCesA10) and cellulose synthase-like (e.g., AtCslB2) genes, not previously implicated in the plant defense response, was altered markedly and differentially by various treatments. Transcripts of some genes required for cell maintenance (e.g., methionine synthase, formate dehydrogenase) and genes involved in sugar metabolism (e.g., glucose transporter), seemed to be required at a higher level after at least one treatment. The induction of genes for sugar transport after microbial challenge and elicitor treatment has been reported previously (36). In addition, significant changes in the expression profiles of other genes with no previous functional description were observed. Of 270 genes with no known function, 106 genes showed significant induction or repression profiles. These included 28, 32, 41, and 8 genes after Alternaria inoculation, SA, MJ and ethylene treatments, respectively, whereas repressed transcript abundances were measured for 2, 14, 18, and 1 genes, respectively.
Analysis of Coordinated Plant Defense Responses.
The analysis of global expression profiles of a large number of Arabidopsis genes in response to infection and treatment with signal molecules provided a base to identify commonalities among defense pathways. A comparison of expression profiles from all four treatments revealed 126 genes (approximately 5%) induced by multiple treatments that are likely to be regulated by the same or overlapping defense signaling pathways (Fig. 2). This may be a conservative estimate considering the stringent measures applied for data analysis. The most significant coinduction pattern was observed for 55 genes that were coinduced by SA and MJ treatments (Fig. 2). This contradicts previous notions that the salicylate and jasmonate pathways might be antagonistic (37). However, our data also revealed eight genes that were significantly induced by SA and significantly repressed by MJ (e.g., germin-like protein, chalcone synthase, and lipid-transfer protein; Fig. 3). This indicates that signal antagonism may be specific to particular genes.
Half of the genes induced by ethylene were also induced by MJ treatment (Fig. 2; e.g., genes of AP2 domain-containing proteins, plant defensins (PDF1.2), an ethylene receptor (ERS2), GAST1 protein homologue, cyclophilin, GA-1, selenium-binding protein; Fig. 3). Overlaps of the MJ and ethylene-signaling pathways have previously been reported for individual genes (e.g., PDF1.2; 7 and EIN2; 11). Ten genes, mostly encoding putative regulatory proteins (e.g., ARR2 and AP2 domain-containing proteins; Fig. 3), were induced by all of the chemical signal treatments. Of 168 genes that were significantly induced by fungal inoculation, 21, 33, and 4 genes (50 genes in total) were also induced by SA, MJ, and ethylene, respectively. The largest number of genes (28 genes) corepressed was again observed for the SA and MJ treatments, including genes with putative regulatory functions (examples in Fig. 3). These results indicate a substantial coregulation or crosstalk among the different plant defense pathways.
Importantly, however, some gene transcripts were induced by one signal molecule and not significantly induced by the others. Some examples of these treatment-specific genes are listed in Fig. 3 and include flavonol synthase, receptor protein kinase, and RNA-binding protein cp29 for SA, catalases, blue copper protein, an isoflavone reductase homologue for MJ, and a protein kinase for the ethylene treatment. Interestingly, expression of some genes was significantly induced by one treatment while also significantly repressed by another treatment. These genes that seem to be highly regulated provide a good starting point for further research to study possible interactions among the components of plant defense signaling pathways.
Our data also suggest the existence of crosstalk between defense and other signaling pathways. Induction of genes encoding chlorophyll A/B-binding proteins (CAB) by the SA treatment is one of the examples of such coordinate responses (Fig. 3). Results presented here further support the hypothesis that the SA-mediated signaling pathway crosstalk with the pathway regulated with the phytochrome A/red light, leading to the induction of CAB genes (38). Availability of an Arabidopsis phytochrome A and B signaling mutant (psi2) showing elevated level of PR gene expression also suggests the notion that light signal transduction and pathogenesis-related gene-signaling pathways are connected (39). In contrast, coordinate response between MJ and light signal pathways resulted in down-regulation of genes encoding ribulose bisphosphate carboxylase (examples in Fig. 3).
In conclusion, high-throughput microarray analysis used in this study permitted the simultaneous analysis of changes that occurred in the transcriptional activities of selected Arabidopsis genes on activation of defense responses. Even though the number of genes analyzed represented only a small selected subset of approximately 24,000 coding sequences, estimated to be present in the complete Arabidopsis genome, large sets of informative data were generated that led to the identification of a high number of potential defense-related transcripts. The results demonstrated that a substantial network of regulatory interactions exists and that considerable interaction occurs among the different defense signaling pathways, notably between the SA and MJ pathways, which were previously believed to act in an antagonistic fashion. Further, more detailed information revealed by simultaneous analyses of large numbers of genes is likely to extend our knowledge of the complicated network of biochemical and regulatory interactions associated with particular biological processes, such as plant defense.
Supplementary Material
Acknowledgments
This research was supported by the Grains Research and Development Corporation (GRDC) of Australia (to J.M.) and by the U. S. Department of Energy and the Carnegie Institution of Washington (to S.S.). We are grateful to Dr. C. Somerville for kind provision of the Arabidopsis EST collection, to Prof. P. Gresshoff, Prof. J. Irwin, Dr. R. Casu, Dr. K. Goulter, and H. Way for critical manuscript reading, and to Dr. D. Maclean, Dr. P. Ebert, R. Brown, and E. Badruzsaufari for useful discussions.
Abbreviations
- ESTs
expressed sequence tags
- MJ
methyl jasmonate
- SA
salicylic acid
References
- 1.Broekaert W F, Cammue B P A, De Bolle M F C, Thevissen K, De Samblanx G V, Osborn R W. Crit Rev Plant Sci. 1997;16:297–323. [Google Scholar]
- 2.Yun D-J, Bressan R, Hasegawa P M. Plant Breeding Rev. 1997;14:39–88. [Google Scholar]
- 3.Grant M, Mansfield J. Curr Opin Plant Biol. 1999;2:312–319. doi: 10.1016/S1369-5266(99)80055-7. [DOI] [PubMed] [Google Scholar]
- 4.Delaney T P, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 5.Cao H, Glazebrook J, Clarke J D, Volko S, Dong X. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 6.Penninckx I A M A, Eggermont K, Terras F R G, Thomma B P H J, De Samblanx G W, Buchala A, Métraux J-P, Manners J M, Broekaert W F. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penninckx I A M A, Thomma B P H J, Buchala A, Métraux J-P, Broekaert W F. Plant Cell. 1998;10:2103–2114. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieterse C M J, van Wees S C M, van Pelt J A, Knoester M, Laan R, Gerrits H, Weisbeek P J, van Loon L C. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomma B P H, Eggermont K, Penninckx I A M A, Mauch-Mani B, Vogelsang R, Cammue B P A, Broekaert W F. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reymond P, Farmer E E. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 12.Alonso J M, Hirayama T, Roman G, Nourizadeh S, Ecker J R. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 13.Eisen M, Spellman P, Brown P, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spellman P, Sherlock G, Zhang M, Iyer V, Anders K, Eisen M, Brown P, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White K P, Rifkin S A, Hurban P, Hogness D S. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 16.Jelinsky S A, Samson L D. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T A. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Höfte H, Desprez T, Amselem J, Chiapello H, Rouze P, Caboche M, Moisan A, Jourjon M F, Charpenteau J L, Berthomieu P, et al. Plant J. 1993;4:1051–1061. doi: 10.1046/j.1365-313x.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- 20.Cooke R, Raynal M, Laudie M, Delseny M. Plant J. 1997;11:1127–1140. doi: 10.1046/j.1365-313x.1997.11051127.x. [DOI] [PubMed] [Google Scholar]
- 21.Newman T, deBruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruan Y, Gilmore J, Conner T. Plant J. 1998;15:821–833. doi: 10.1046/j.1365-313x.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- 23.Heller R A, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley D E, Davis R W. Proc Natl Acad Sci USA. 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–257. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 25.Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. Plant Cell. 1999;11:431–444. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarre D A, Wolpert T J. Plant Cell. 1999;11:237–249. doi: 10.1105/tpc.11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu X, Reddy A S. Plant Mol Biol. 1997;34:949–959. doi: 10.1023/a:1005893119263. [DOI] [PubMed] [Google Scholar]
- 28.Dong X, Mindrinos M, Davis K R, Ausubel F M. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richmond T, Somerville S. Curr Opin Plant Biol. 2000;3:108–116. doi: 10.1016/s1369-5266(99)00049-7. [DOI] [PubMed] [Google Scholar]
- 30.Riechmann J L, Meyerowitz E M. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- 31.Menke F L H, Champion A, Kijne J W, Memelink J. EMBO J. 1999;18:4455–4463. doi: 10.1093/emboj/18.16.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melan M A, Dong X, Endara M E, Davis K R, Ausubel F M, Peterman T K. Plant Physiol. 1993;101:441–450. doi: 10.1104/pp.101.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietrich R A, Richberg M H, Schmidt R, Dean C, Dangl J L. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S, Klessig D F. Plant Cell. 1997;9:809–824. doi: 10.1105/tpc.9.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conrath U, Silva H, Klessig D F. Plant J. 1997;11:747–757. [Google Scholar]
- 36.Truernit E, Schmid J, Epple P, Illig J, Sauer N. Plant Cell. 1996;8:2169–2182. doi: 10.1105/tpc.8.12.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doares S H, Narvaes-Vasquez J, Conconi A, Ryan C A. Plant Physiol. 1995;108:1741–1746. doi: 10.1104/pp.108.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genoud T, Métraux J-P. Trends Plant Sci. 1999;4:503–507. doi: 10.1016/s1360-1385(99)01498-3. [DOI] [PubMed] [Google Scholar]
- 39.Genoud T, Millar A J, Nishizawa N, Kay S A, Schafer E, Nagatani A, Chua N H. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.