Abstract
Negative transcriptional regulation or cross-coupling between NF-κB (RelA) and the glucocorticoid receptor (GR) is proposed to play a regulatory role in human physiology and disease. Despite previous advances, the biochemical basis of this phenomenon remains a subject of controversy. We show here that the inhibition of GR activity by RelA does not require the RelA DNA binding, transactivation, or nuclear localization domains. Surprisingly, RelA repression of GR is abolished by mutation of the conserved protein kinase A (PKA) site at amino acid residue 276 of RelA. We show that GR associates in vivo and in vitro with the catalytic subunit of PKA (PKAc) in a ligand-independent manner and that GR transcription depends on PKA signaling. Indeed, we demonstrated that GR-mediated inhibition of NF-κB transactivation is PKAc-dependent. In contrast to previous models, we suggest that the cross-coupling requires a cytoplasmic step and is regulated by a PKAc-associated signaling.
Signaling from cell surface receptors to the nucleus is transmitted by the translocation of activated protein kinases and/or activated transcription factors from the cytoplasmic to the nuclear compartment (1). Lipophilic ligands, such as the steroids and retinoids, directly bind to their intracellular cognate nuclear hormone receptors so that no second message is required for transmitting the signals (2–6). Although nuclear receptor-dependent transcriptional activation relies on receptor and its DNA binding element interactions, nuclear receptors are able to modulate the activity of other classes of trans-activators through DNA-binding independent mechanism(s). This process of regulation is known as transcriptional cross-talk or cross-coupling (5, 7).
In the last several years, the biochemical basis of transcriptional cross-talk has been under intense investigation. For example, the glucocorticoid receptor (GR), estrogen receptor, retinoic acid receptor, and the thyroid hormone receptor have been shown to “cross-couple” with AP-1, E26 transformation-specific-related proteins, and NF-κB (7–12). Inhibition of the proinflammatory factor, NF-κB, by activated GR has been suggested to play a key role in GR-mediated anti-inflammatory responses (12–14).
Direct protein–protein interaction between cross-coupled transcription factors has been suggested to be one mechanism of inhibition (5, 15–20). It also has been suggested that cross-talk is mediated by direct competition for the coactivator cAMP response element-binding protein-binding protein/p300 (21, 22). To date, all cross-talk mechanisms have been proposed to occur in the nucleus.
In unstimulated cells, both the GR and NF-κB are predominantly sequestered in the cytoplasm. Cytoplasmic retention of GR involves binding of unliganded GR to hsp90 and other regulatory proteins whereas that for NF-κB involves inhibitor proteins, such as IκBα. Nuclear translocation of GR is induced by association with the natural glucocorticoids or the synthetic analogs such as dexamethasone (Dex) (2, 23, 24). In contrast, NF-κB translocation is associated with stimulation of cells with a variety of agents, such as lipopolysaccharide, tumor necrosis factor-α (TNF-α), and IL-1, environmental stresses, or the Tax protein of the type 1 human T cell leukemia virus, which induce degradation of the inhibitor protein IκB (12, 25–27).
Besides the physical partitioning of inactive NF-κB to the cytosol, it recently has been shown that NF-κB transcription is regulated through phosphorylation of RelA by protein kinase A (PKA) (28). The catalytic subunit of PKA (PKAc) associates with RelA in the cytoplasm promoting RelA phosphorylation at Ser-276. NF-κB activators induce IκBα degradation, enabling translocation of the phosphorylated RelA to the nucleus, where it binds to its cognate DNA response elements and enhances transcription (28).
PKA exerts a regulatory role in the activation of multiple nuclear hormone receptors. For example, it has been shown that PKAc activates GR-dependent DNA binding in cotransfection studies (29). In addition, PKAc can directly phosphorylate GR in vitro (30), enhances transcription by the retinoic acid receptor (31, 32), and regulates dimerization of human estrogen receptor-α (33). cAMP activators potentiate ligand-dependent and -independent transcription by the thyroid or retinoid-x receptor and the progesterone receptors, respectively (34, 35). At physiologic levels glucocorticoids and cAMP synergistically promote apoptotic cell death (36). Even though the exact biochemical function of activated PKAc in the presence or absence of steroids and/or retinoids remains unclear, it is apparent that PKA may represent an important regulatory mechanism in nuclear hormone receptor signaling.
We now demonstrate that PKAc associates with GR and potentiates GR-dependent transcription. We show that PKAc attenuates NF-κB and GR cross-repression. Mutation of p65 at the conserved PKA phosphorylation site, amino acid 276, abolishes the potential of p65 to repress GR. Our results localize the NF-κB and GR cross-coupling to the cytoplasm and implicate PKAc-dependent signaling as the molecular interface of this mutual inhibition.
Materials and Methods
Antibodies and Reagents.
Purified polyclonal rabbit antiserum against human GR (GR-135 rabbit polyclonal antibody) and the rabbit RelA antibody 5314 have been described (37). The RelA goat antibody (G20), the antibodies for IκBα, and the rabbit GR (P-20) antibody (sc-1002) were purchased from Santa Cruz Biotechnology. The PKAc mouse mAb (P73420) was from Transduction Laboratories, Lexington, KY. Secondary antibodies labeled either with FITC or Texas red were purchased from The Jackson Laboratory. Recombinant human TNF-α (10 ng/ml), H-89 (10–40 mM) isoquinoline-sulfonamide derivative, which is one of the most potent inhibitors of PKA [Ki (inhibition constant) of ≈0.048 mM], and ML-7 (40 mM) inhibitor specific for myosin light chain kinase were purchased from Calbiochem, and 8-Br cAMP (0.1 mM), dexamethasone (0.01–1 μM) and 12-O-tetra-decanoyl-phorbol 13-acetate (TPA) (100–200 ng/ml) were from Sigma. All reagents were used at the indicated concentrations and applied in <0.1% of the media volume.
The mouse mammary tumor virus (MMTV)-luciferase (Luc), Igk3-tk-Luc, Rous sarcoma virus (RSV)GR, RSVI550, RSVGRΔ589–697, and CMXGR and CMXβgal expression vectors have been described (24, 38, 39). The cytomegalovirus (CMV) IκBα and the human CMVRelA have been described (25). RelA mutants were kindly provided by D. W. Ballard, Pennsylvania State University College of Medicine (40). CMVRelA, CMVRelAΔNLS (nuclear localization vector), and CMVRelA31–551 express the corresponding p65-coding sequence downstream of the CMV promoter. Glutathione-S-transferase expression vector (pGEX) PKAc and pcPKAc were kindly provided by S. Taylor, University of California at San Diego.
Cell Culture.
CV1, MEF, 293, and HeLaS3 cells were maintained as monolayers in DMEM supplemented with 100 units/ml penicillin–streptomycin and 10% resin-charcoal-stripped bovine calf serum (38) or 10% heat-inactivated FCS, respectively (GIBCO). HeLa cells were transfected at low serum concentration (0.1%). Cultures were maintained at 37°C and in 7% CO2. For immunofluorescence, cells were grown on round coverslips (Corning Glass) in 6-well plates.
Transfection and Reporter Assays.
Transfection assays were done as described (38). Luc and β-galactosidase were assayed as described (39). Cell extracts were prepared 24–30 h after transfection or as indicated in the figure legends. Results are given as a relative activity, based on the positive control activity (arbitrary units) as observed in each described experiment.
In Vitro Binding Assays.
Binding assays were done as described (38). In summary, glutathione S-transferase (GST)-PKAc was prepared as indicated (Amersham Pharmacia). Radio-labeled RelA wild-type and mutant proteins and GR protein were prepared by coupled in vitro transcription-translation (Promega) by using the corresponding expression vector as template DNA. For in vitro binding assays, 30–50 μl of glutathione Sepharose associated with the corresponding recombinant protein was incubated with 3–5 μl of 35S-labeled proteins, in the presence or absence of dexamethasone, for 30 min at 4°C. The bound proteins were washed five times in appropriate buffers and analyzed by SDS/PAGE and autoradiography.
Immunohistochemistry Assays.
Cells were fixed and analyzed by immunofluorescence studies as described (41). Fluorescence images were analyzed by confocal microscopy. For in vitro immunodetection assay, total cytoplasmic, nuclear, or whole-cell extracts (WCE) were analyzed in SDS/PAGE, transferred to nitrocellulose membrane, and probed with the corresponding antibodies as described (37). Immunoprecipitations were carried out by using the indicated specific antibodies as described (42).
Results
GR Represses NF-κB-Mediated Transcription.
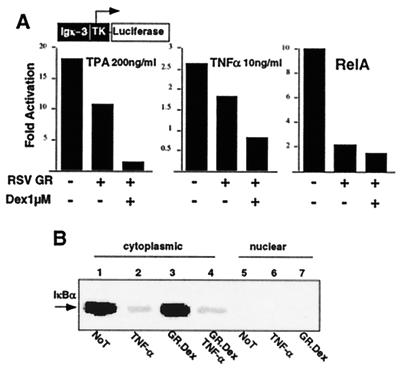
Fig. 1A shows that both TPA and TNF-α activates NF-κB transcriptional activity. Overexpression of wild-type GR protein represses NF-κB-dependent transcription. Addition of Dex leads to further repression of NF-κB activation (Fig. 1A and data not shown). These data suggest that GR represses NF-κB activity in both ligand-dependent and -independent manners. Ligand independent NF-κB repression also has been reported to be mediated by another member of the steroid receptor family, the progesterone receptor (18).
Figure 1.
GR mediates trans-repression of TNF-α and TPA-activated RelA-dependent signaling. (A) GR represses NF-κB-dependent transcription. CV1 cells (48-well plates) were transiently cotransfected with 120 ng of Igk3-Luc reporter construct, 75 ng of CMXβgal, and 75 ng of RSVGR or RSV empty expression vectors. Cells were costimulated with TPA or TNF-α in the absence or presence of Dex for 4 h before the assay, as indicated. The y axis shows activation as measured by Igk3-driven Luc activity. The histogram is representative of three independent experiments. (B) Protein levels of IkBα are not affected by GR.Dex treatment in 293 cells. Western blot analysis of IkBα in cytoplasmic or nuclear extracts in cells without treatment (Not), treated with TNF-α, overexpression of GR and Dex treatment (GR.Dex), or in combination of TNF-α and GR.Dex.
It has been suggested previously that inhibition of NF-κB by GR might involve Dex-dependent transcriptional activation of the IκBα gene to increase IκBα protein, which, in turn, inhibits nuclear translocation of NF-κB (13, 14). To determine the absolute requirement of IκBα in GR-mediated NF-κB repression, we examined IκBα protein levels upon Dex treatment. Western blot analysis of IκBα protein from cytoplasmic and nuclear cell extracts revealed that GR/Dex neither up-regulated IκBα protein nor affected TNF-α mediated IκBα degradation (Fig. 1B). This result suggests that GR/Dex neither up-regulated IκBα at the protein levels nor affected TNF-α-mediated IκBα degradation.
Cytoplasmic-Localized RelA Represses GR Activity.
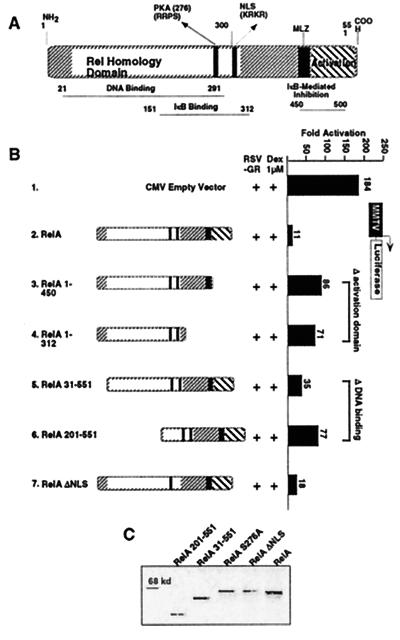
To further investigate the cross-coupling phenomenon, we analyzed the domains of RelA and GR proteins involved in these interactions. RelA is characterized by an N-terminal conserved Rel homology domain (which contains regions required for DNA binding, dimerization, nuclear localization, and PKAc-mediated phosphorylation) and a C-terminal transactivation domain (Fig. 2A). The wild-type RelA protein transfected at a 1:3 M ratio with GR repressed MMTV activation >15-fold (Fig. 2B, compare constructs 1 and 2). The RelA mutants missing the activation domain (RelA 1–450 and RelA 1–312; Fig. 2B, constructs 3 and 4) or the DNA-binding domain (RelA 31–551 and RelA 201–551; Fig. 2B, constructs 5 and 6) were still able to repress GR transcription, although the repression was reduced by 7- to 8-fold. The expression levels of both wild-type and mutant proteins are shown in Fig. 2C. These results suggest that the RelA DNA binding and activation domains play a role in repression of GR activity. On the other hand, although the extent of GR inhibition by these RelA mutants is less than that induced by the wild-type protein, these results also indicate that repression of GR can still occur even in the absence of RelA DNA binding and trans-activating properties.
Figure 2.
Mapping the domain of RelA involved in GR repression. (A) Schematic of RelA. (B) RelA cross-coupling domains. CV1 cells (48-well plates) were transiently cotransfected with 120 ng of MMTV-Luc reporter construct, 75 ng of CMXβgal, 37.5 ng of RSVGR, and CMV empty vector or CMVRelA mutants at 3:1 M ratio with GR, as indicated. Cells were stimulated with Dex at 1 μM for 10 h before the assay. The y axis shows activation as measured by MMTV-driven Luc activity conducted in quadruplicate assays. (C) Expression levels of RelA proteins. Western blot analysis of RelA wild-type and mutant proteins.
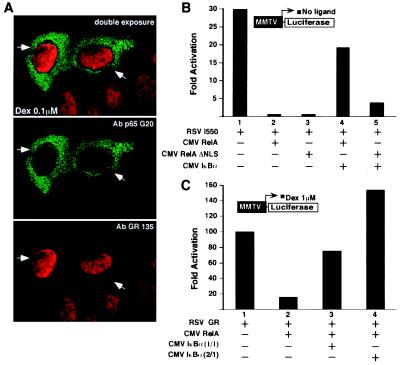
We next analyzed the effect of RelAΔNLS on GR-mediated transcription. Deletion of the nuclear localization signal (RelAΔNLS) results in a predominantly cytoplasmic protein (40, 43). Surprisingly, RelAΔNLS blocks hormone-dependent GR transcription at levels equivalent to the wild-type RelA (Fig. 2B, construct 7). Immunofluorescence studies from transfected cells with RelAΔNLS expression vector confirmed a virtually complete cytoplasmic distribution of RelAΔNLS protein (Fig. 3A). Furthermore, RelA and RelAΔNLS repress equally well the constitutively active and constitutively nuclear localized GR mutant (I550) (Fig. 3B, lanes 1–3). Considering that GR I550 carries a deleted ligand-binding domain, we propose that neither the ligand-binding domain nor its associated activation function (AF-2) is necessary for the RelA inhibition. In parallel experiments, cotransfection of IκBα reversed the p65-mediated repression of GR I550 in a concentration-dependent manner (Fig. 3B, compare lanes 2 and 4; data not shown). As expected, the ability of RelAΔNLS to block GR was only partially blocked by IκBα overexpression (Fig. 3B, lanes 3 and 5), in agreement with the poor IκBα-RelAΔNLS association efficiency (40, 43).
Figure 3.
Cytoplasmic form of RelA controls GR transcription. (A) Immunohistochemistry detecting relative cellular distribution of Dex-activated GR and RelAΔNLS proteins. Confocal image represents a double-exposure photograph. Primary antibodies are used as indicated. Green corresponds to the RelA labeling revealed with the FITC-conjugated second antibody, and red corresponds to the GR labeling revealed with the Texas red conjugated second antibody. HeLa cells were transiently cotransfected with the CMVRelAΔNLS expression vector and the MMTV-Luc reporter, as indicated in Fig. 2B. Cells were treated with 0.1 μM Dex for 3 h before fixation. The filled arrowheads point to the transfected cells. Similar results were obtained in CV1 cells by using ectopically expressed GR and RelAΔNLS proteins (data not shown). (B) IκBα-free RelA wild type and RelAΔNLS equally repress a constitutively active GR mutant (I550). The transient transfection assay was performed as in Fig. 2B. The RSVI550 was used at 37.5 ng, and the CMVIκBα was transfected at 1:1 M ratio with the RelA expression vector. All of the transfections were equalized for the total amount of expression vectors, i.e., CMV empty vector. (C) IκBα sequesters RelA and blocks RelA-mediated GR repression. The experiment was performed as above but cells were stimulated with Dex at 1 μM for 10 h before the assay. The CMVIκBα was used at 1:1 or 1:2 M ratio with RelA expression vector, as indicated. The y axis shows activation as measured by MMTV-driven Luc activity conducted in quadruplicate assays.
A Mutation of PKA Site in RelA Abolishes the Repression of GR Activity.
Because the p65 DNA binding, transactivation, NLS, and dimerization domains are not required for inhibition of GR activity, we examined the role of the conserved PKAc phosphorylation site at position 276 (Fig. 2A) in repression. We tested two phosphorylation negative RelA mutants for their ability to affect GR activation. Surprisingly, both serine to alanine or serine to glycine substitutions (Ser-276–Ala and Ser-276–Gly) completely abolished GR repression (Fig. 4, lanes 1–4). This result suggests that, in Dex-treated cells, GR-dependent transcriptional activation is severely repressed by a cytoplasmic IκBα-free RelA protein and this effect is inactivated either by IκBα overexpression (Fig. 3C) or RelA residue 276 mutation (Fig. 4). These results identified Ser-276 as a critical residue for the RelA-mediated GR repression and suggest that PKAc-dependent phosphorylation of RelA may represent a key biochemical interface of RelA and GR cross-talk.
Figure 4.
Point mutations of RelA PKA phosphorylation site abolish RelA-mediated GR repression. A conserved PKA phosphorylation site of RelA, Ser-276, was mutated to Ala (S276A) or Gly (S276G). Transfection was performed as in Fig. 2B. CMV RelA mutants were assayed as indicated. The y axis shows activation as measured by MMTV-driven Luc activity conducted in quadruplicate assays.
GR Associates with PKAc in Vitro and in Vivo.
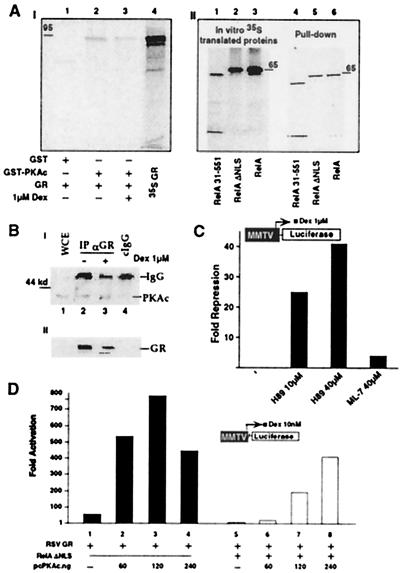
In vitro association studies show that GR physically interacts with PKAc (Fig. 5A). A series of pull-down experiments with in vitro-translated GR and RelA proteins and the bacterially produced GST-PKAc fusion were carried out. In the presence and absence of Dex, ≈30% of input wild-type GR was pulled down with GST-PKAc after a 30-min incubation (Fig. 5AI, lanes 1–4). In control experiments, the three major repression-active RelA proteins, wild-type RelA., RelAΔNLS, and RelA31–551 were equally pulled down by the GST-PKAc fusion (Fig. 5AII, lanes 1–6).
Figure 5.
PKAc is associated with GR and potentiates GR activity. (A) In vitro association of PKAc subunit and GR (95 kDa). (AI) The 35S-labeled GR protein was incubated with GST-PKAc or GST alone on glutathione-agarose beads, in the absence or presence of 1 μM Dex as indicated. The bound proteins were analyzed by SDS/PAGE and fluorography (lanes 1–3). Input was included in lane 4. (AII) The same conditions as in AI but using 35S-labeled RelA wild-type and mutants. Input proteins (lanes 1–3) and the pull-down proteins (lanes 4–6) are indicated. (B) In vivo association of PKAc and GR wild-type protein. (BI) Human 293 cells were cotransfected with CMXGR and pcPKAc expression vectors. Cells were mock-treated (lane 2) or activated with 1 μM Dex for 12 h (lane 3) and harvested 48 h after transfection. Cell lysates were immunoprecipitated with 5 μl of GR-135 polyclonal antibody (lanes 2 and 3) or with the same amount of normal rabbit IgG as a control (lane 4). Immunoprecipitates then were analyzed by Western blot with PKAc-specific antibody. WCE were loaded in parallel with the immunoprecipitates to show comigration (lane 1). (BII) An aliquot of the WCE was analyzed directly with Western blot by using GR-specific antibody. (C) H-89, a PKAc-specific inhibitor represses GR activity. CV-1 cells were transfected with MMTV-Luc, CMXβgal, and RSVGR as in Fig. 2B. After transfection cells were stimulated for 4 h with 1 μM Dex, with or without H-89 or ML-7, at the indicated concentrations. Cellular extracts were used to measure Luc activity. The y axis shows arbitrary unit of repression as measured by MMTV-driven Luc activity. The histogram is representative of at least three independent experiments. (D) PKAc potentiates GR transcription and reverses RelA repression of GR. CV1 cells were cotransfected with 120 ng of MMTV-Luc reporter construct, 75 ng of CMXβgal, 33 ng of RSVGR, CMV empty vector or CMVRelA ΔNLS at 120 ng, and increasing concentration of PKAc expression vector, as indicated. Cells were stimulated with Dex at 10 nM for 10 h before the assay. All of the transfections were normalized for the total amount of expression vectors, i.e., CMV empty vector. The y axis shows arbitrary unit of activation as measured by MMTV-driven Luc activity.
At the in vivo level, human 293 cells were cotransfected with the expression vectors encoding for GR and PKAc at 1:1 M ratio (Fig. 5B). Transfected cells were lysed, and immunoprecipitations were performed with antibody against GR (Fig. 5BI). The immunoprecipitates were analyzed by SDS/PAGE and immunoblotted with an antibody specific for PKAc. The PKAc protein could be coprecipitated with GR by using GR-specific antibody, but not with control IgG, indicating the in vivo presence of a PKAc-GR complex formation (Fig. 5BI, compare lanes 2 and 4). In control experiments, similar amounts of WCE were immunoblotted with GR antibody and data confirmed the expected expression of overexpressed GR protein (Fig. 5AII). In agreement with the transfected data, PKAc was immunoprecipitated with GR from HeLa S3 cells, further indicating the in vivo presence of a functional GR/PKAc complex (unpublished data).
PKA Activates GR-Mediated Transcription.
The in vitro and in vivo data supporting the GR/PKAc association suggested that PKA-mediated phopshorylation might modulate GR transcription. To elucidate the possible role of PKA signaling on GR transactivation, we used the isoquinoline-sulfonamide derivative, H-89, one of the most potent inhibitors of PKA [Ki (inhibition constant) of ≈0.048 mM]. As a control, we used ML-7, another inhibitor that is specific for myosin light chain kinase. We tested the effect of the two inhibitors on Dex-activated GR-dependent transcription, using a transient transfection assay based on expression of Luc from a reporter construct, MMTV-Luc. We observed an important inhibition of Dex-induced GR-dependent transcription in the presence of 10–40 μM H-89 (Fig. 5C). In contrast, ML-7, when used at up to 40 μM, had much less effect on GR-dependent transcription (Fig. 5C). Taken in consideration that H-89 did not affect GR nuclear translocation (data not shown), this experiment provides strong evidence for the involvement of PKA in modulating GR-mediated transcriptional activation.
Taken together, the above data suggested that inactivation or sequesteration of PKA signaling could be the source of the repression of GR activity. To test this competition model, we monitored GR transcription in the presence of 10 nM Dex and increasing amounts of PKAc expression vector (Fig. 5D). In the absence of overexpressed PKAc, GR activated MMTV transcription 60-fold, whereas expression of PKAc stimulated GR an additional 10- to 15-fold (Fig. 5D, lanes 1–4). In the presence of RelAΔNLS (3:1 M ratio with GR) MMTV transcription was severely repressed (Fig. 5D, compare lanes 1 and 5). However, high-level expression of PKAc rescued GR activity to 400-fold (Fig. 5D, compare lanes 4, 5, and 8). These data suggest that the concentration of PKAc in the cell may be critical in controlling GR transactivation and RelA-mediated GR repression.
GR Represses PKAc-Potentiated NF-κB Activity.
To follow the role of PKAc concentration in the cross-talk of GR-RelA, we performed a PKAc-sequestration assay in vivo by modifying the molecular equilibrium between RelA and GR proteins. Human 293 cells were cotransfected with the expression vectors for RelA and PKAc at 1:1 M ratio, along with empty or GR expression vectors (Fig. 6AI). The immunoprecipitates were analyzed by SDS/PAGE and immunoblotted with antibody specific for PKAc. Indeed, PKAc is associated with RelA in the absence of overexpressed GR protein (Fig. 6AI). However, overexpression of GR in the absence or presence of Dex strongly interferes with the PKAc-RelA interaction, resulting in decreased association of RelA and PKAc in vivo (Fig. 6A I and II). As expected, the control experiments confirmed the overexpression of GR and RelA proteins from WCE (Fig. 6A III and IV). Considering the essential role of PKAc in RelA-mediated transactivation (28), the above result suggests that overexpression of GR should interfere with PKAc-mediated RelA transactivation.
Figure 6.
GR competes with RelA for PKAc association in vivo. (A) GR blocks RelA:PKAc protein association in vivo. Human 293T cells were transfected with 5 μg of CMVRelA, 5 μg of pcPKAc, and 5 μg of CMXGR or empty vector, as indicated. Cells were mock-treated or activated with 1 μM Dex for 12 h and harvested 48 h after transfection. (AI) Cell lysates were immunoprecipitated with 5 μl (1 μg) of RelA sc-109 rabbit polyclonal antibody (lanes 3–6) or with the same amount of normal rabbit IgG as control (lane 2). Immunoprecipitates were analyzed by Western blot with PKAc-specific antibody. WCE (lane 1) were loaded in parallel with the immunoprecipitates to show comigration. (AII) Quantitation of PKAc coprecipitated by RelA-specific antibody. The y axis shows the ratio of PKAc to IgG. Quantitation is performed with NIH image program. (AIII) Similar amount of WCE from cells corresponding to lanes 3–6 in AI was immunostained with GR-specific antibody (sc-1002). (AIV) The same blot shown in AIII was stripped and reprobed with RelA-specific antibody. (B) GR represses PKAc-dependent NF-κB transactivation. CV1 cells were cotransfected with 120 ng of Igk3-Luc reporter construct, 75 ng of CMXβgal, 33 ng of CMV RelA, and increasing amount of pcPKAc expression vector, as indicated. The RSVGR and RSVGRΔ589–697 were used at 3:1 M ratio with RelA. All of the transfection points were equalized for the total amount of expression vectors. The y axis shows fold activation as measured by Igk3-driven Luc activity. The histogram is representative of a triplicate experiment.
To further test the above hypothesis, we monitored NF-κB transcription in the presence of 33 ng of RelA expression vector and increasing amounts of PKAc (Fig. 6B). In the absence of PKAc, RelA activated NF-κB reporter (Igk3-Luc) arbitrarily 1-fold, whereas expression of PKAc stimulated NF-κB transcription an additional 12-fold (Fig. 6B, lanes 1–4). In the presence of wild-type GR or GRΔ589–697, a mutant GR protein carrying a truncated ligand-binding domain and characterized by a predominant cytoplasmic localization (24), NF-κB transcription was severely repressed (Fig. 6B, lanes 1–4). This result demonstrates that GR could compete with RelA for PKAc and represses PKAc-mediated NF-κB transcription.
Discussion
In this study, we have re-examined the biochemical basis of the functional cross-repression between NF-κB and the GR. Two previously described pathways for NF-κB and GR cross-talk proposed either a direct physical interaction resulting in the mutual loss of DNA-binding activity of each factor (13, 15, 17–20) or competition for a limiting common transcriptional coactivator (21, 22). A third mechanism involves hormone induced synthesis of IκBα in the repression of NF-κB by GR, which might be a cell type-specific event (13, 14). Our study suggests that the trans-repression of GR and RelA depends on a cytoplasmic event that requires both PKAc and the PKA phosphorylation site in RelA.
In characterizing the repression, we found that the RelA mutants with deletion of either the DNA-binding domain or the activation domain remain repressive to GR activity, although to a less extent than that of the wild-type RelA. However, the cytoplasmic RelAΔNLS mutant retained full repression activity, indicating that a cytoplasmic event is involved in the cross-repression. Furthermore, a mutant GR (GRΔ589–697), characterized by a predominant cytoplasmic localization, is able to repress RelA-dependent transcription, further suggesting that the cross-repression of GR and RelA is mediated by a cytoplasmic process. This study introduces a unique concept for the cross-talk between transcription factors because all previous cross-talk mechanisms implicated a nuclear process.
The observation that RelA mutated at the conserved PKA-phosphorylation site completely lost the capacity to inhibit GR directly implicated PKA in this cross-talk. Traditionally, PKA activation requires cAMP, which binds to the regulatory subunit of the PKA complex and releases the catalytic subunit of PKAc (44, 45). However, a recent study by Ghosh and coworkers (28) demonstrated that some PKAc exists as part of an inactive complex with NF-κB/IκB. Cytokine activation promotes degradation of IκBα or IκBβ, which activates translocation of a PKAc-phosphorylated and transcriptionally active RelA to the nucleus. Our observations that expression of PKAc restores GR activity despite the presence of RelA, together with the results on competition of PKAc association between GR and RelA, implicates PKA as a potential source of the repression. Indeed, we demonstrated that GR/RelA cross-repression could be relieved by overexpression of PKAc. These results together suggest that PKAc is involved in the GR–NF-κB cross-talk pathway.
Another observation presented in this paper is that GR represses RelA-activated transcription in both ligand-dependent and -independent manner. Previous studies have shown that treatment of cells with Dex, a synthetic GR ligand, leads to repression of NF-κB-activated transcription. In this study, we demonstrated that overexpression of GR results in the inhibition of both TPA/TNFα-activated or RelA-overexpression-mediated transcription in a ligand-independent manner. Addition of GR ligand leads to a further repression of NF-κB activity. This study suggests that multiple layers of regulation are involved in the transrepresison of GR and RelA.
More interestingly, the cross-talk involving PKAc is not restricted to GR/NF-κB. A similar PKAc-dependent mechanism contributes to transrepression by NF-κB and other nuclear receptors such as retinoic acid receptor (data not shown). PKAc potentiates all-trans retinoic acid-activated transcription, whereas RelA represses all-trans retinoic acid-induced transcription. Coexpression of PKAc with RelA relieves repression of retinoic acid receptor-target gene expression by RelA. The data together suggest that the PKAc signaling might serve as a general component for trans-repression of nuclear receptors and NF-κB.
Even though our data suggested that PKA signaling might act upstream of Dex-activated GR transcription, the mechanism by which PKAc controls GR activity is not clear. Possibly, PKA signaling might promote a change in the tertiary structure of the GR, which affects affinity of GR for transcriptional coregulators. This is an attractive hypothesis because ligand activation of the nuclear receptors is possibly related to a conformational change that favors coactivator interaction (46). An alternative possibility is that PKA has a more direct role by altering receptor interaction with transcriptional cofactors, as has been observed for the interaction between phosphorylated cAMP response element-binding protein and its main coactivator CBP (38, 47). The concept that GR and possibly other nuclear receptor functioning is regulated by a cytoplasmic PKA-associated signaling as has been described for the NF-κB pathway, provides a completely new framework for understanding how the cross-talk integrates a variety of signals in a physiological context. Further elucidation of such a mechanism would not only reveal how cross-talk controls signal activated transcription but also provide an approach to study the transcriptional basis of anti-inflammatory, antiviral, and anticancer agents.
Acknowledgments
This paper is dedicated to the memory of our friend and colleague Dr. Kazuhiko Umesono. We thank S. Taylor, D. W. Ballard, and D. Baltimore for reagents, B. Seufzer for excellent technical assistance, and P. Olson and W. Xu for critical reading of the manuscripts. A.W. was a research assistant at the Salk Institute, S.M. is an assistant professor at the University of Wisconsin Madison Medical School, I.V. is a professor of the American Cancer Society at the Salk Institute, V.D. is a maître Assistant at the University of Geneva Medical School, and R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by University of Geneva Medical School, by University of Wisconsin Madison Medical School, by a Shaw Scientist Award (S.M.), by Howard Hughes Medical Institute, and in part by National Institutes of Health Grants GM26444 and HD27183.
Abbreviations
- GR
glucocorticoid receptor
- PKAc
catalytic subunit of protein kinase A
- CMV
cytomegalovirus
- WCE
whole-cell extracts
- Dex
dexamethasone
- GST
glutathione S-transferase
- RSV
Rous sarcoma virus
- CMX
cytomegalovirus expression vector
- TPA
12-O-tetra-decanoyl-phorbol 13-acetate
- NLS
nuclear localization signal
- TNF-α
tumor necrosis factor-α
- MMTV
mouse mammary tumor virus
- Luc
luciferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220413297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220413297
References
- 1.Karin M, Hunter T. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 2.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 5.Beato M, Herrlich P, Schutz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 6.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 7.Schule R, Rangarajan P, Kliewer S, Ransone L J, Bolado J, Yang N, Verma I M, Evans R M. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- 8.Yang-Yen H F, Chambard J C, Sun Y L, Smeal T, Schmidt T J, Drouin J, Karin M. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 9.Schule R, Rangarajan P, Yang N, Kliewer S, Ransone L J, Bolado J, Verma I M, Evans R M. Proc Natl Acad Sci USA. 1991;88:6092–6096. doi: 10.1073/pnas.88.14.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucas V, Spyrou G, Yaniv M. EMBO J. 1991;10:2237–2245. doi: 10.1002/j.1460-2075.1991.tb07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauthier J M, Bourachot B, Doucas V, Yaniv M, Moreau-Gachelin F. EMBO J. 1993;12:5089–5096. doi: 10.1002/j.1460-2075.1993.tb06203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 13.Scheinman R I, Cogswell P C, Lofquist A K, Baldwin A S., Jr Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- 14.Auphan N, DiDonato J A, Rosette C, Helmberg A, Karin M. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 15.Ray A, Prefontaine K E. Proc Natl Acad Sci USA. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheinman R I, Gualberto A, Jewell C M, Cidlowski J A, Baldwin A S., Jr Mol Cell Biol. 1995b;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein B, Yang M X. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalkhoven E, Wissink S, van der Saag P T, van der Burg B. J Biol Chem. 1996;271:6217–6224. doi: 10.1074/jbc.271.11.6217. [DOI] [PubMed] [Google Scholar]
- 19.Palvimo J J, Reinikainen P, Ikonen T, Kallio P J, Moilanen A, Janne O A. J Biol Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- 20.Wissink S, van Heerde E C, Schmitz M L, Kalkhoven E, van der Burg B, Baeuerle P A, van der Saag P T. J Biol Chem. 1997;272:22278–22284. doi: 10.1074/jbc.272.35.22278. [DOI] [PubMed] [Google Scholar]
- 21.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 22.Sheppard K A, Phelps K M, Williams A J, Thanos D, Glass C K, Rosenfeld M G, Gerritsen M E, Collins T. J Biol Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- 23.Evans R M. Recent Prog Horm Res. 1989;45:1–22. doi: 10.1016/b978-0-12-571145-6.50005-4. ; discussion 22–27. [DOI] [PubMed] [Google Scholar]
- 24.Cadepond F, Gasc J M, Delahaye F, Jibard N, Schweizer-Groyer G, Segard-Maurel I, Evans R, Baulieu E E. Exp Cell Res. 1992;201:99–108. doi: 10.1016/0014-4827(92)90352-9. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto S, Verma I M. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 26.Baldwin A S., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 27.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 29.Rangarajan P N, Umesono K, Evans R M. Mol Endocrinol. 1992;6:1451–1457. doi: 10.1210/mend.6.9.1435789. [DOI] [PubMed] [Google Scholar]
- 30.Haske T, Nakao M, Moudgil V K. Mol Cell Biochem. 1994;132:163–171. doi: 10.1007/BF00926925. [DOI] [PubMed] [Google Scholar]
- 31.Huggenvik J I, Collard M W, Kim Y W, Sharma R P. Mol Endocrinol. 1993;7:543–550. doi: 10.1210/mend.7.4.8388997. [DOI] [PubMed] [Google Scholar]
- 32.Rochette-Egly C, Oulad-Abdelghani M, Staub A, Pfister V, Scheuer I, Chambon P, Gaub M P. Mol Endocrinol. 1995;9:860–871. doi: 10.1210/mend.9.7.7476969. [DOI] [PubMed] [Google Scholar]
- 33.Chen D, Pace P E, Coombes R C, Ali S. Mol Cell Biol. 1999;19:1002–1015. doi: 10.1128/mcb.19.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitman D C, Costa C H, Graf H, Baxter J D, Ribeiro R C. J Biol Chem. 1996;271:21950–21955. doi: 10.1074/jbc.271.36.21950. [DOI] [PubMed] [Google Scholar]
- 35.Denner L A, Weigel N L, Maxwell B L, Schrader W T, O'Malley B W. Science. 1990;250:1740–1743. doi: 10.1126/science.2176746. [DOI] [PubMed] [Google Scholar]
- 36.Gruol D J, Altschmied J. Mol Endocrinol. 1993;7:104–113. doi: 10.1210/mend.7.1.8383286. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto S, Maki M, Schmitt M J, Hatanaka M, Verma I M. Proc Natl Acad Sci USA. 1994;91:12740–12744. doi: 10.1073/pnas.91.26.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doucas V, Egan D A, Tini M, Evans R M. Proc Natl Acad Sci USA. 1999;96:2627–2632. doi: 10.1073/pnas.96.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doucas V, Evans R M. Proc Natl Acad Sci USA. 1999;96:2633–2638. doi: 10.1073/pnas.96.6.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganchi P A, Sun S C, Greene W C, Ballard D W. Mol Biol Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Mosser D D, Morimoto R I. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 44.Taylor S S, Buechler J A, Yonemoto W. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 45.Scott J D. Pharmacol Ther. 1991;50:123–145. doi: 10.1016/0163-7258(91)90075-w. [DOI] [PubMed] [Google Scholar]
- 46.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 47.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]