Abstract
Demonstrating in vivo interaction of two important biomolecules and the relevance of the interaction to a biological process have been difficult issues in biomedical research. Here, we report the use of a homology modeling approach to establish the significance of protein interactions in governing the activation of programmed cell death in Caenorhabditis elegans. A protein interaction cascade has been postulated to mediate activation of cell death in nematodes, in which the BH3-domain-containing (Bcl-2 homology region 3) protein EGL-1 binds the cell-death inhibitor CED-9 and induces release of the death-activating protein CED-4 from inhibitory CED-4/CED-9 complexes. We show here that an unusual gain-of-function mutation in ced-9 (substitution of glycine 169 to glutamate) that results in potent inhibition of most nematode cell deaths impairs the binding of EGL-1 to CED-9 and EGL-1-induced release of CED-4 from CED-4/CED-9 complexes. Based on a modeled EGL-1/CED-9 complex structure, we generated second-site compensatory mutations in EGL-1 that partially restore the binding of EGL-1 to CED-9(G169E) and EGL-1-induced release of CED-4 from CED-4/CED-9(G169E) complexes. Importantly, these mutations also significantly suppress the death-protective activity of CED-9(G169E) in vivo. These results establish that direct physical interaction between EGL-1 and CED-9 is essential for the release of CED-4 and the activation of cell death. The structure-based design of second-site suppressors via homology modeling should be widely applicable for probing important molecular interactions that are implicated in fundamental biological processes.
Programmed cell death is a tightly regulated cellular process crucial for metazoan development and homeostasis (1, 2). Improper regulation of programmed cell death can lead to a variety of diseases, including cancer and degenerative disorders (3). Genetic analysis of programmed cell death in Caenorhabditis elegans has identified four genes whose activities are essential for proper activation and execution of programmed cell death (4). Three of them promote cell death (ced-3, ced-4, and egl-1), and the fourth, ced-9, protects against cell death (5–7). Importantly, these genes encode proteins that share significant sequence and functional homology with mammalian cell death regulators. EGL-1 is similar to BH3-only pro-apoptotic proteins (7, 8), CED-3 is a member of the aspartate-specific cysteine protease family (caspases) (9, 10), CED-4 is similar to one of the apoptotic protease-activating factors (Apaf-1) (11, 12), and CED-9 is a member of a family of anti-apoptotic proteins first defined by the mammalian Bcl-2 protein (8, 13, 14). Some of the C. elegans proteins and their mammalian homologues have been shown to be functionally interchangeable (4), indicating that the cell death pathway is evolutionarily conserved.
Bcl-2 was first identified as an inhibitor of apoptosis by virtue of its ability to protect against lymphocyte cell death (14–16). Subsequently, a family of Bcl-2-related proteins, which have at least one of the four conserved BH domains, was identified (8, 14), some of which (e.g., Bak and EGL-1) are pro-apoptotic, whereas others (e.g., Bcl-2 and CED-9) are anti-apoptotic. Several biochemical properties have been associated with Bcl-2 family proteins, including the ability to localize to outer membranes of mitochondria and nuclei and membranes of endoplasmic reticulum, to form heterodimers with other family members, and to form ion-conducting channels in synthetic membranes (reviewed in ref. 17). However, the precise functions associated with each biochemical property remain poorly understood, especially the function of heterodimerization between Bcl-2-like proteins and proteins bearing an amphipathic helical BH3 domain (17). Study of the biochemical functions of EGL-1 and CED-9, the invertebrate prototypes of BH3-only proteins and Bcl-2-like proteins, may provide important insights into this fundamental question.
In C. elegans, CED-9 localizes to mitochondria and is capable of interacting with both EGL-1 and CED-4 in various in vitro protein interaction assays (18 and reviewed in ref. 19). CED-4 colocalizes with CED-9 at mitochondria in living cells but translocates to nuclear membranes in cells induced to die by overexpression of EGL-1, suggesting that CED-9 and CED-4 may physically associate at mitochondria in vivo and that EGL-1 may cause cell death by releasing CED-4 from CED-4/CED-9 complexes (18). These observations lead to a protein interaction cascade model in which binding of EGL-1 to CED-9 induces release of CED-4 from mitochondria-localized CED-4/CED-9 complexes and subsequent translocation of CED-4 to nuclear membranes. Intriguingly, translocation of CED-4 to nuclear membranes is blocked by an unusual gain-of-function mutation (n1950) in the ced-9 gene that potently inhibits almost all programmed cell deaths in C. elegans and is a result of substitution of glycine 169 with glutamate in the BH1 domain of CED-9 (18, 20). This finding suggests that CED-9(G169E) either is unable to interact with EGL-1 or is unable to release CED-4. However, no difference has been detected between the interactions of EGL-1 with CED-9 or CED-9(G169E) by in vitro protein interaction assays (18). This result raises important questions regarding whether EGL-1 interacts directly with CED-9 in vivo or indirectly through an intermediate protein to release CED-4 and the mechanistic basis of n1950 in blocking the translocation of CED-4 from mitochondria to nuclear membranes.
In this report, we carried out biochemical analyses to demonstrate that the ced-9(n1950) mutation blocks release of CED-4 from CED-4/CED-9 complexes by impairing the binding of EGL-1 to CED-9, providing a mechanistic explanation for the function of the ced-9(n1950) mutation. Furthermore, using homology modeling, we constructed a modeled structure of the EGL-1/CED-9 complex and used it to design and generate second-site compensatory mutations in EGL-1 that suppress the activities of CED-9(G169E) both in vitro and in vivo, providing convincing evidence that EGL-1 and CED-9 interact physically in C. elegans and that such interaction is essential for activating cell death in nematodes.
Materials and Methods
Plasmid Construction.
We used standard methods of molecular biology to generate various EGL-1, CED-9, and CED-4 constructs and their mutant derivatives (21).
Protein Purification.
Glutathione S-transferase (GST)-CED-4 was expressed in Escherichia coli XA-90 cells and purified by glutathione affinity chromatography as instructed by the manufacturer (Amersham Pharmacia). His-tagged CED-9 proteins were expressed in E. coli BL21(DE3)pLysS cells and purified by Ni-NTA affinity chromatography as directed by the supplier (Qiagen). GST-CED-4 and His6CED-9(68–251) or His6CED-9(68–251; G169E) were coexpressed in BL21(DE3)pLysS cells and copurified by either glutathione or Ni-NTA affinity chromatography. EGL-1 proteins were expressed in BL21(DE3)pLysS and purified from inclusion bodies using the method for purification of UNC-86 (22). EGL-1 proteins prepared using this procedure are about 90% pure.
Electrophoretic Mobility-Shift Assays.
Purified CED-9 was incubated with purified EGL-1 in CED-3 buffer (23) containing 2.5 mM DTT/0.01% Nonidet P-40/10% glycerol at room temperature (unless otherwise indicated) for 30 min, resolved on 6% native polyacrylamide gels (PAGE) containing 0.01% Nonidet P-40 at 4°C, and then transferred to nitrocellulose membrane for Western blotting and ECL detection (Amersham Pharmacia). Nonidet P-40 was added to binding reactions where indicated.
CED-4 Release Assays.
GST-CED-4/His6CED-9(68–251) complexes (wild type or G169E) were purified and immobilized on glutathione-Sepharose beads, and 500 ng of EGL-1 protein was added to the immobilized protein complexes in the presence of 0.25% Nonidet P-40. Supernatants were collected after 30 min at room temperature and resolved by native PAGE. The EGL-1/CED-9 complexes released were detected by Western blot analysis using anti-His6 antibodies. Alternatively, protein complexes were eluted from glutathione-Sepharose with 20 mM reduced glutathione, immobilized on Ni-NTA agarose, and subsequently incubated with 500 ng of EGL-1 protein as described above. Supernatants were analyzed by SDS-PAGE, and the release of GST-CED-4 was detected by Western blotting using anti-GST antibodies.
Structural Modeling.
Homology modeling of the structure of the CED-9/EGL-1 BH3 domain complex was performed with the SWISS-MODEL package (24) using the published structure of the Bcl-xL/Bak BH3 domain complex as a template (25). The modeled CED-9/EGL-1 structure was further evaluated by packing density analysis and residue profiling using SWISS-MODEL and judged to be reasonable.
Results and Discussion
The G169E Substitution in CED-9 Impairs the Binding of EGL-1 to CED-9 but Does Not Affect Association of CED-9 with CED-4.
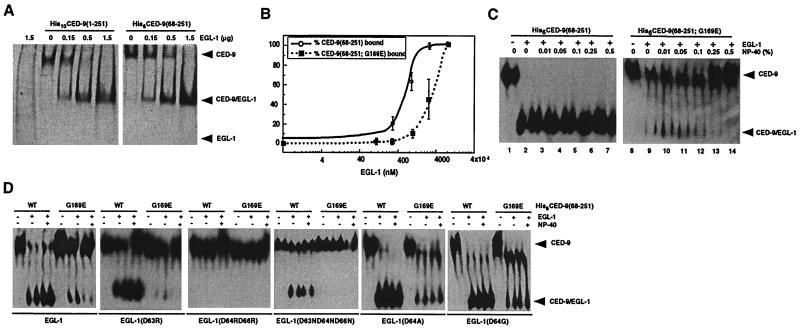
Because the n1950 mutation in CED-9 alters a highly conserved residue (Gly-169) in the BH1 domain, which has been implicated in mediating protein interactions (26), we investigated whether this mutation affects the binding of EGL-1 to CED-9 or formation of CED-4/CED-9 complexes. Using deletion analysis and limited protease digestion analysis, we identified a core domain of CED-9 (amino acids 68 to 251) that is sufficient to mediate interactions of CED-9 with both EGL-1 and CED-4 (Figs. 1A and 2A, and data not shown). This core domain is also sufficient to confer the gain-of-function phenotype associated with CED-9 (G169E) (ref. 27 and data not shown). This protease-resistant core () was thus used for the in vitro protein interaction assays described below. Using electrophoretic mobility-shift assays, which resolve proteins or protein complexes by native PAGE, we found that the G169E substitution in CED-9 reduced the binding affinity of EGL-1 for CED-9 by about 4-fold in the absence of nonionic detergent (Fig, 1B). In the presence of nonionic detergent (0.1% or more Nonidet P-40), the affinity of EGL-1 for CED-9(68–251; G169E) was further reduced, whereas the binding of EGL-1 to CED-9(68–251) was unaffected (Fig. 1C). These results suggest that the G169E mutation impairs the binding of EGL-1 to CED-9. The observation that nonionic detergent destabilizes binding of EGL-1 to CED-9(68–251; G169E) but not CED-9(68–251) may have significant implications regarding the interaction of EGL-1 and CED-9 in vivo. Both EGL-1 and CED-9 may localize at, or insert into, membrane-bound organelles such as mitochondria (18) in a fashion analogous to their mammalian counterparts, Bid and Bcl-xL, both of which contain structural motifs similar to those of the membrane translocation domains of bacterial toxins, such as diphtheria toxin (28–32).
Figure 1.
Characterization of interactions between EGL-1 and CED-9 proteins. (A) CED-9(68–251) is sufficient to bind EGL-1. Purified His10CED-9(1–251) or His6CED-9(68–251) (1.5 μg each) was incubated with the indicated amount of EGL-1 and subjected to native PAGE followed by Coomassie blue staining. (B) The G169E substitution in CED-9 impairs the binding of EGL-1 to CED-9. Purified His6CED-9(68–251) or His6CED-9(68–251; G169E) (500 ng each) was incubated with increasing concentrations of purified EGL-1 and then subjected to native PAGE and Western blotting analysis using anti-His6 antibodies. The amounts of bound and unbound CED-9 were quantified using ImageQuant software (Molecular Dynamics). The percentage of CED-9 in complex with EGL-1 is displayed as a function of the amount of EGL-1 used in the binding reactions. (C) Formation of CED-9(G169E)/EGL-1 complexes but not CED-9/EGL-1 complexes is sensitive to the concentration of the non-ionic detergent Nonidet P-40. Binding reactions were carried out as described (B) using 500 ng of CED-9 and EGL-1 proteins in the presence of the indicated amount of Nonidet P-40. (D) Interactions between CED-9 and mutant EGL-1 proteins. Wild-type or mutant EGL-1 proteins (250 ng each) were incubated with 500 ng of the indicated CED-9 proteins in the presence or absence of 0.25% Nonidet P-40, and the assays were carried out as described (B).
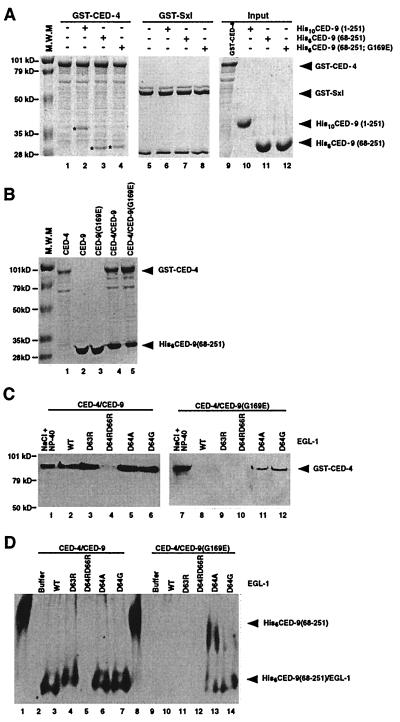
Figure 2.
Interactions of CED-9 with CED-4 and EGL-1-induced release of CED-4 from CED-4/CED-9 complexes. (A) His6CED-9(68–251; G169E) binds GST-CED-4 as well as His6CED-9(68–251). Purified CED-9 proteins (2.5 μg) were incubated with an equivalent amount of GST-CED-4 or GST-Sxl immobilized on glutathione-Sepharose beads. Protein complexes were washed three times with CED-3 buffer (23) containing 0.01% Triton X-100 and subjected to SDS-PAGE followed by Coomassie Blue staining. The CED-9 proteins pulled down by GST-CED-4 are indicated with asterisks (lanes 2–4). (B) CED-4 copurifies with CED-9 proteins in 1:1 ratios. GST-CED-4 and His6CED-9(68–251) or His6CED-9(68–251; G169E) were expressed alone or were coexpressed in E. coli and purified using affinity chromatography. (C) EGL-1 releases CED-4 from CED-4/CED-9 complexes. Five-hundred nanograms of wild-type or mutant EGL-1 was added to approximately 1 μg of purified GST-CED-4/His6CED-9(68–251) or GST-CED-4/His6CED-9(68–251; G169E) complexes immobilized on Ni-NTA beads, and the resulting supernatants were subjected to SDS-PAGE and Western blot analysis using anti-GST antibodies to assess the amount of GST-CED-4 released. As positive controls, 1 M NaCl and 1% Nonidet P-40 were used to disassociate GST-CED-4 from GST-CED-4/CED-9 complexes. (D) EGL-1 displaces CED-4 from CED-4/CED-9 complexes. Five-hundred nanograms of wild-type or mutant EGL-1 was added to approximately 1 μg of purified GST-CED-4/His6CED-9(68–251) or GST-CED-4/His6CED-9(68–251; G169E) complexes immobilized on glutathione-Sepharose beads, and the resulting supernatants were subjected to native PAGE and Western blot analysis using anti-His6 antibodies to visualize the amount of EGL-1/CED-9 complexes released. In lanes 1 and 8, purified CED-9 (250 ng) was loaded as a control for free CED-9 species. EGL-1/CED-9 complexes containing EGL-1(D63R), EGL-1(D64A), or EGL-1(D64G) migrate slower than complexes with wild-type EGL-1 as a result of loss of a negatively charged Asp residue.
We found that the G169E substitution in CED-9 did not seem to affect the binding of CED-4 to CED-9 based on two different experiments. First, in GST-fusion protein pull-down experiments, GST-CED-4 bound CED-9(68–251) and CED-9(68–251; G169E) with comparable affinities (Fig. 2A, lanes 3 and 4). Similar results were obtained when different concentrations of CED-9 proteins (wild type or G169E) were incubated with GST-CED-4 or when a different CED-4 fusion protein (CED-4-FLAG) was used in pull-down experiments using anti-FLAG affinity chromatography (data not shown). Second, when we coexpressed GST-CED-4 with CED-9(68–251) or CED-9(68–251; G169E) in bacteria, we found that both CED-9(68–251) and CED-9(68–251; G169E) copurified with GST-CED-4 in approximate 1:1 ratios, further indicating that the G169E substitution does not affect binding of CED-4 to CED-9 (Fig. 2B, lanes 4 and 5).
EGL-1 Induces Release of CED-4 from CED-9/CED-4 Complexes but Not from CED-9(G169E)/CED-4 Complexes.
We next tested whether the reduced binding of EGL-1 to CED-9(G169E) affected release of CED-4 from CED-4/CED-9(G169E) complexes. We examined this process using two different assays. In the first assay, we monitored EGL-1-induced release of GST-CED-4 from GST-CED-4/His6CED-9(68–251) or GST-CED-4/His6CED-9(68–251; G169E) complexes immobilized on Ni-NTA beads that bound the six-histidine tag of the CED-9 proteins. We found that GST-CED-4 was released from GST-CED-4/His6CED-9(68–251) complexes but not from GST-CED-4/His6CED-9(68–251; G169E) complexes by addition of EGL-1 (Fig. 2C, lanes 2 and 8). In the second assay, we analyzed both the release of CED-9 from GST-CED-4/CED-9 complexes immobilized on glutathione-Sepharose beads, which bound GST-CED-4 and the formation of EGL-1/CED-9 complexes using the electrophoretic mobility-shift assay. As shown in Fig. 2D (lanes 3 and 10), upon addition of EGL-1, His6CED-9(68–251) was released from GST-CED-4/His6CED-9(68–251) complexes, and new EGL-1/His6CED-9(68–281) complexes were formed, indicative of the displacement of CED-4 from CED-4/CED-9 complexes by EGL-1. In contrast, release of His6CED-9(68–251; G169E) from GST-CED-4/His6CED-9(68–251; G169E) complexes was undetectable. Taken together, these results suggest that the G169E substitution in CED-9 impairs the binding of EGL-1 to CED-9 and the ability of EGL-1 to release CED-4 from CED-4/CED-9 complexes, resulting in enhanced death-protective activity of CED-9(G169E) in C. elegans. This finding provides a mechanistic explanation for the interesting observation that the n1950 mutation blocks the translocation of CED-4 from mitochondria to nuclear membranes induced by expression of EGL-1 (18). Furthermore, our ability to reconstitute the early death-activation events (EGL-1-mediated release of CED-4) in vitro using purified recombinant proteins strongly suggests that the proposed protein interactions are direct.
Homology Modeling of the CED-9/EGL-1 Complex Structure and Structure-Based Design of Second-Site Suppressors of the ced-9(n1950) Mutation.
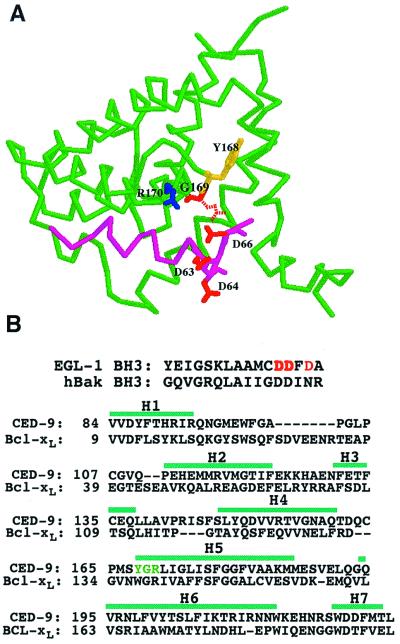
To understand the structural basis of the effect of the G169E substitution on the binding of EGL-1 to CED-9, we carried out homology modeling of the structure of the complex between the EGL-1 BH3 domain and CED-9 based on the published structure of the complex between Bcl-xL, a CED-9 homologue, and a 16-amino acid BH3 domain peptide from the pro-apoptotic protein Bak (25). In this modeled structure, Gly-169 of CED-9 sits at the base of a hydrophobic cleft formed by the BH1, BH2, and BH3 domains of CED-9 and may be in close contact with one or more of four residues (Asp-63, Asp-64, Phe-65, and Asp-66) at the carboxyl terminus of the BH3 domain of EGL-1 (Fig. 3A). Substitution of glutamate for Gly-169 may destabilize the interaction between EGL-1 and CED-9 by two possible mechanisms. First, the negatively charged glutamate in CED-9(G169E) may electrostatically repulse one or more of the three negatively charged residues in the BH3 domain of EGL-1 (Asp-63, Asp-64, and Asp-66). Alternatively, the bulkier glutamate side chain may block the access of the BH3 domain of EGL-1 to the hydrophobic binding pocket in CED-9. To distinguish between these two possibilities, we attempted to make compensatory second-site mutations in EGL-1 that might alleviate either the electrostatic repulsion (D63R, D64RD66R, or D63ND64ND66N) or steric hindrance (D64G or D64A) caused by the G169E substitution in CED-9. We found that none of the EGL-1 proteins carrying the first set of mutations (D63R, D64RD66R, or D63ND64ND66N) bound CED-9(68–251; G169E) (Fig. 1D). These mutations also affect the binding of EGL-1 to CED-9(68–251); the binding affinity of EGL-1(D63R) or EGL-1(D63ND64ND66N) for CED-9(68–251) was partially reduced, and the binding of EGL-1(D64RD66R) to CED-9(68–251) was totally abolished (Fig. 1D). In contrast, both EGL-1 mutant proteins of the second class (D64G or D64A), while binding CED-9(68–251) as well as EGL-1, bound CED-9(68–251; G169E) significantly better than EGL-1 and formed stable complexes that are not sensitive to treatment with Nonidet P-40 (Fig. 1D). These results suggest that steric hindrance caused by the G169E substitution in CED-9 can partially account for the reduced binding of EGL-1 to CED-9(68–251; G169E), but they do not rule out the possibility that electrostatic repulsion may still play a role in destabilizing the interaction of EGL-1 with CED-9(68–251; G169E).
Figure 3.
Modeled structure of the complex between the BH3 domain of EGL-1 and CED-9. (A) Ribbon stereodrawing of the modeled complex. Backbones of CED-9 and EGL-1 (BH3 domain) are shown in green and magenta, respectively. Potentially critical interface residues are depicted with colored sticks: Y168 (yellow), G169 (red), and R170 (blue) in CED-9 and D63, D64, and D66 (all in red) in EGL-1. The dashed Glu residue at the position of G169 in CED-9 indicates the gain-of-function mutation in CED-9. (B) Sequence alignments between the BH3 domains of EGL-1 and human Bak and between CED-9 and Bcl-xL. The sequence alignment between CED-9 and Bcl-xL was optimized using the SWISS-MODEL program (24). The previously defined seven α-helices (H1–H7) in Bcl-xL are indicated with green bars (25).
Consistent with the hypothesis that the binding of EGL-1 to CED-9 is important for the release of CED-4 from CED-4/CED-9 complexes, none of the EGL-1 proteins carrying the first set of mutations was capable of promoting release of CED-4 from CED-4/CED-9(68–251; G169E) complexes, whereas both EGL-1(D64G) and EGL-1(D64A) were able to displace a significant amount of CED-4 from CED-4/CED-9(68–251; G169E) complexes by forming new complexes with CED-9(68–251; G169E) (Fig. 2C, lanes 7–12; Fig. 2D, lanes 8–14). Reinforcing this conclusion, EGL-1 mutants that retained the ability to bind CED-9 (D63R, D64A, or D64G) were also able to release CED-4 from CED-4/CED-9(68–251) complexes, whereas EGL-1(D64RD66R), which did not bind CED-9, failed to release CED-4 (Fig. 1D; Fig. 2C, lanes 3–6; Fig. 2D, lanes 4–7).
Second-Site Mutations in EGL-1 Suppress the ced-9(n1950) Mutation in Vivo.
We next examined whether the ability of EGL-1 to bind CED-9 and to release CED-4 from CED-4/CED-9 complexes in vitro correlates with its ability to induce cell death in nematodes. Briefly, we generated transgenic nematodes expressing either wild-type or mutant EGL-1 proteins under the control of C. elegans heat-shock promoters or the endogenous egl-1 promoter. We then determined the extent of cell killing induced by expression of these proteins by counting the number of cell corpses generated in embryos of transgenic animals (7). The transgenes were introduced into ced-1(e1735); egl-1(n3082) animals to test for rescue of the egl-1(n3082) loss-of-function phenotype and ced-1(e1735); ced-9(n1950) animals to test for cell killing in the ced-9(n1950) background (the ced-1 mutation results in a persistent cell corpse phenotype, facilitating the scoring of cells that have undergone programmed cell death; ref. 33). When examined at 20°C, late-stage embryos of ced-1(e1735), ced-1(e1735); egl-1(n3082), and ced-1(e1735); ced-9(n1950) animals have an average of 31, 0.2, and 3.3 cell corpses, respectively, indicating that in the absence of EGL-1 or in the presence of CED-9(G169E) very few cell deaths occur (Table 1). Heat shock-induced expression of the wild-type EGL-1 protein restored cell killing and resulted in an average of around 40 cell corpses in ced-1(e1735); egl-1(n3082) animals but failed to induce any additional cell death in ced-1(e1735); ced-9(n1950) animals (Table 1). In contrast, expression of the EGL-1(D64RD66R) protein, which could not bind either CED-9 or CED-9(G169E), did not induce any additional cell death in either of the mutant animals (Fig. 1D and Table 1). Furthermore, expression of either EGL-1(D63R) or EGL-1(D63ND64ND66N), both of which bind CED-9 but not CED-9(G169E), induced robust cell killing in ced-1(e1735); egl-1(n3082) animals but did not cause any additional cell death in ced-1(e1735); ced-9(n1950) animals (Fig. 1D and Table 1). These results provide further support for the hypothesis that the binding of EGL-1 to CED-9 is important for its cell-killing activity.
Table 1.
The cell-killing activities of EGL-1 and EGL-1 mutants
| hsp construct* | Array |
egl-1(n3082)
|
ced-9(n1950)
|
||
|---|---|---|---|---|---|
| No. of embryos | No. of corpses† | No. of embryos | No. of corpses† | ||
| None | 30 | 0.2 ± 0.4 | 30 | 3.3 ± 1.8 | |
| Vector | 1 | 30 | 0.3 ± 0.5 | 30 | 1.6 ± 1.6 |
| 2 | 30 | 0.2 ± 0.5 | 30 | 2.7 ± 1.7 | |
| 3 | 30 | 0.3 ± 0.5 | 30 | 2.6 ± 1.3 | |
| EGL-1 | 1 | 31 | 46.0 ± 24.6 | 30 | 4.0 ± 1.9 |
| 2 | 31 | 33.6 ± 20.9 | 30 | 3.8 ± 1.8 | |
| 3 | 30 | 40.5 ± 15.7 | 30 | 2.3 ± 1.8 | |
| EGL-1(D63R) | 1 | 32 | 43.4 ± 24.3 | 31 | 2.6 ± 2.3 |
| 2 | 30 | 31.9 ± 25.1 | 30 | 2.8 ± 2.7 | |
| 3 | 30 | 46.9 ± 13.2 | 30 | 3.2 ± 2.0 | |
| EGL-1(D64RD66R) | 1 | 30 | 0.6 ± 1.3 | 30 | 3.2 ± 2.6 |
| 2 | 33 | 0.2 ± 0.6 | 32 | 2.6 ± 1.5 | |
| 3 | 31 | 0.7 ± 1.3 | 30 | 3.4 ± 1.8 | |
| EGL-1(D63ND64ND66N) | 1 | 30 | 34.8 ± 17.0 | 30 | 3.6 ± 2.5 |
| 2 | 30 | 32.2 ± 19.0 | 30 | 2.6 ± 1.5 | |
| 3 | 30 | 35.0 ± 17.6 | 30 | 3.4 ± 1.9 | |
The hsp constructs (at 50 μg/ml each) were injected into ced-1(e1735); egl-1(n1084 n3082) unc-76(e911) animals with pTG96 (at 20 μg/ml), which expresses GFP in many somatic cells in most of the developmental stages (35), and p76-16B (at 50 μg/ml), which rescues unc-76 uncordinated phenotype (36). The transgenes were then crossed into ced-1(e1735); ced-9(n1950) mutant background.
Heat shock experiments and cell-killing assays were performed as described (7). Fluorescent transgenic embryos at the three-fold or later stages of embryogenesis were scored for cell corpses in the head region. Data shown are means ± SEM. All data except the first row depicts results obtained after heat-shock treatment. For comparison, ced-1(e1375) animals have an average of about 31 cell corpses in the same region.
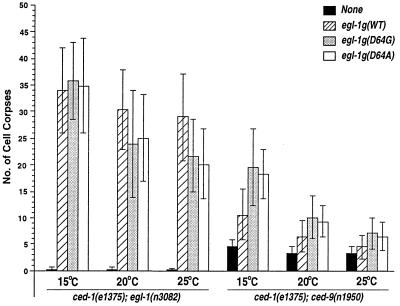
Interestingly, when EGL-1(D64A) or EGL-1(D64G) proteins, which bind CED-9(G169E) significantly better than EGL-1 and bind CED-9 as well as EGL-1, were expressed under the control of heat-shock promoters, we observed inconsistent cell killing in both ced-1(e1735); egl-1(n3082) and ced-1(e1735); ced-9(n1950) animals, ranging from robust cell killing to no cell killing (data not shown). One possible explanation for this inconsistency in cell killing, which was not seen with other EGL-1 mutant proteins, is that the mutations (D64A or D64G) may have temperature-sensitive effects on either the stability of EGL-1 or its interaction with CED-9. Indeed, we found that the in vitro binding of both EGL-1(D64G) and EGL-1(D64A) to CED-9 (wild type or G169E) was destabilized at 33°C, the temperature used for the heat-shock experiments (data not shown). The binding of EGL-1 to CED-9 or CED-9(G169E) was also affected by temperature, but to a much lesser extent (data not shown; see below). To examine the death-inducing activity of EGL-1(D64G) and EGL-1(D64A) proteins at normal temperatures, we expressed these mutant proteins under the control of the promoter of egl-1 gene, which is thought to be expressed in most cells destined to die in nematodes (34), and scored the cell-killing activity of the mutant proteins at three temperatures (15, 20, and 25°C) normally used to grow nematodes. As described above, few cell deaths occurred in ced-9(n1950) animals. An average of 4.5, 3.3, and 3.2 cell corpses was observed in ced-1(e1735); ced-9(n1950) animals at 15, 20, and 25°C, respectively (Fig. 4). Expression of the wild-type EGL-1 under the control of the egl-1 promoter resulted in increased numbers of cell deaths in ced-1(e1735); ced-9(n1950) animals at 25, 20, and 15°C by an average of one, three, and six cell corpses, respectively, indicating that the CED-9(G169E) mutant responds to higher levels of EGL-1 expression in a temperature-sensitive manner (Fig. 4). Consistent with these in vivo observations, we found that EGL-1 binds CED-9(G169E) in vitro with higher affinity at 15°C than at 25°C (data not shown). Importantly, at all three temperatures tested, both EGL-1(D64G) and EGL-1(D64A) mutant proteins clearly demonstrated stronger death-inducing activity in ced-1(e1735); ced-9(n1950) animals than the wild-type EGL-1 protein (P < 0.0001), with the strongest death-inducing activity observed at 15°C [an average of 20 cell corpses was observed with EGL-1(D64G); Fig. 4]. These results indicate that the D64G and D64A substitutions, which enhance the binding of EGL-1 to CED-9(G169E), also significantly enhance the cell-killing activity of EGL-1 in ced-9(n1950) mutants. In parallel with these experiments, we examined the death-inducing activity of these two EGL-1 mutant proteins in ced-1(e1735); egl-1(n3082) animals with the wild-type ced-9 gene. We found that expression of EGL-1(D64G) or EGL-1(D64A) under the control of the egl-1 promoter induced robust cell killing at 15°C at a level comparable to that of wild-type EGL-1. This finding is consistent with the results that EGL-1(D64G) and EGL-1(D64A) bound CED-9 and released CED-4 from CED-4/CED-9 complexes in vitro as well as EGL-1 (Figs. 1D, 2C and D, and 4). At 20 and 25°C, the death-inducing activity of EGL-1(D64G) and EGL-1(D64A) was reduced significantly, further confirming the temperature-sensitive nature of these mutations. In contrast, wild-type EGL-1 proteins were mildly temperature-sensitive and induced comparable amounts of cell death at all three temperatures tested (Fig. 4). Taken together, these results provide a strict correlation between the in vivo cell-killing activity of EGL-1 and its binding affinity for CED-9 as well as its ability to release CED-4 from CED-4/CED-9 complexes in vitro, clearly establishing that direct physical interaction between EGL-1 and CED-9 is required in vivo for EGL-1 to induce cell death.
Figure 4.
EGL-1(D64G) and EGL-1(D64A) induce stronger cell killing in ced-9(n1950) mutants than EGL-1. A 10-kb egl-1 wild-type genomic fragment (hatched box) containing 3.8 kb upstream of the egl-1 start codon and 5.7 kb downstream of the egl-1 stop codon (34) or the corresponding egl-1 genomic fragment carrying either the D64G (gray box) or D64A (empty box) mutation was introduced at 40 μg/ml into ced-1(e1735); egl-1(n1084 n3082) unc-76(e911) or ced-1(e1735); ced-9(n1950) animals with pTG96 (20 μg/ml) (35) and p76–16B (50 μg/ml) (36). The transgenic animals were cultured at three temperatures (15, 20, or 25°C), and the cell-killing activity of the EGL-1 proteins was assessed by counting the number of cell corpses in the head region of 3-fold or later-stage transgenic embryos. All data are averages ± standard deviations of results (n > 50) obtained from three independent transgenic lines.
Critical Residues in the BH3 Domain of EGL-1 Differ from Those of Other BH3 Domain-Containing Pro-Apoptotic Proteins.
Asp-63 is a residue in the BH3 domain of EGL-1 that is absolutely conserved in all BH3 domain-containing proteins and has been shown to be critical for heterodimer formation among Bcl-2 family proteins and for the cell-killing activity of BH3 domain-containing pro-apoptotic proteins (reviewed in ref. 8). Interestingly, the EGL-1(D63R) mutant in which Asp-63 was replaced by an oppositely charged arginine was still capable of binding CED-9, albeit with reduced binding affinity, and could still induce efficient cell killing in ced-1(e1735); egl-1(n3082) animals (Fig. 1D and Table 1). Similar results were obtained with another EGL-1 mutant, EGL-1(D63ND64ND66N). In contrast, similar amino acid substitutions at Asp-64 and Asp-66 of EGL-1 (D64RD66R) abolished the binding of EGL-1 to CED-9 and the in vivo killing activity of EGL-1 (Fig. 1D and Table 1). These results and the result that the D64G or D64A substitution can partially restore the binding of EGL-1 to CED-9(G169E) suggest that the less conserved Asp-64 residue, rather than the highly conserved Asp-63 residue, is the critical amino acid at the interface of EGL-1/CED-9 complexes that contributes to the binding specificity of EGL-1 for CED-9. In this regard, EGL-1/CED-9 complexes seem to differ from the human Bcl-xL/Bak complexes used as a template for our homology modeling. One interpretation that is consistent with our results is that the EGL-1 BH3 helix is rotated slightly within the binding pocket in CED-9 when compared with the orientation of the Bak BH3 helix within the Bcl-xL binding pocket.
It is interesting that a similar mutation (G145E) in Bcl-2 does not result in a gain-of-function phenotype (26). It is likely that the interface between Bcl-2 and its cognate BH3-containing ligand is different from that of EGL-1/CED-9. Thus, it will be interesting to see whether the binding specificity among other Bcl-2 family members is determined primarily by conserved or nonconserved residues at the interfaces of the complexes. Our success in generating second-site suppressor mutations based on this cross-species homology modeling should encourage further applications of this approach to investigate molecular interactions such as protein–protein, protein–DNA, and protein–RNA interactions that are crucial for other fundamental biological processes, especially in systems where genetic approaches are not available.
Our biochemical, structural, and functional analyses of the nature of the ced-9(n1950) mutation and the identification of its compensatory suppressor mutations in CED-9 binding partner EGL-1 have established an in vivo mechanism of protein–protein interactions that govern the appropriate activation of programmed cell death in C. elegans. Given that the cell death pathway is highly conserved, the basic components of this regulatory circuit, direct physical interaction between an anti-apoptotic Bcl-2-like death regulator (CED-9) and a BH3-bearing pro-apoptotic protein (EGL-1) resulting in release and activation of another death-activating protein (CED-4) from an inhibitory protein complex (CED-4/CED-9) or from a restricted compartment like mitochondria, could function in regulation of apoptosis in a wide range of species, including humans.
Acknowledgments
We thank N. Ahn, M. Han, and members of the Xue lab and Chen lab for discussions concerning the manuscript; H.R. Horvitz for providing strains and reagents; H. Banerjee and R. Singh for GST-Sxl. J.P. was supported by a National Science Foundation Graduate Research Fellowship. L.C. was supported by a Damon Runyun Scholar Award. D.X. was supported by grants from the Cancer League of Colorado, American Cancer Society, and National Institutes of Health and is a recipient of a Burroughs Wellcome Fund Career Award and the Searle Scholar Award.
Abbreviation
- GST
glutathione S-transferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210391597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210391597
References
- 1.Ellis R E, Yuan J Y, Horvitz H R. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 2.Steller H. Science. 1995;267:1445–1149. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 3.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 4.Horvitz H R. Cancer Res. 1999;59:1701–1706. [PubMed] [Google Scholar]
- 5.Ellis H M, Horvitz H R. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 6.Hengartner M O, Ellis R E, Horvitz H R. Nature (London) 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 7.Conradt B, Horvitz H R. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 8.Kelekar A, Thompson C B. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 9.Yuan J, Shaham S, Ledoux S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 10.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 11.Yuan J, Horvitz H R. Development. 1992;116:309–320. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 12.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 13.Hengartner M O, Horvitz H R. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 14.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 15.Vaux D L, Cory S, Adams J M. Nature (London) 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 16.Nunez G, London L, Hockenbery D, Alexander M, McKearn J P, Korsmeyer S J. J Immunol. 1990;144:3602–3610. [PubMed] [Google Scholar]
- 17.Vander Heiden M G, Thompson C B. Nat Cell Biol. 1999;1:209–216. doi: 10.1038/70237. [DOI] [PubMed] [Google Scholar]
- 18.Chen F, Hersh B M, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz H R. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 19.Hengartner M. Science. 1998;281:1298–1299. doi: 10.1126/science.281.5381.1298. [DOI] [PubMed] [Google Scholar]
- 20.Hengartner M O, Horvitz H R. Nature (London) 1994;369:318–320. doi: 10.1038/369318a0. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Xue D, Tu Y, Chalfie M. Science. 1993;261:1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]
- 23.Xue D, Horvitz H R. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 24.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 25.Sattler M, Liang H, Nettesheim D, Meadows R P, Harlan J E, Eberstadt M, Yoon H S, Shuker S B, Chang B S, Minn A J, et al. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 26.Yin X M, Oltvai Z N, Korsmeyer S J. Nature (London) 1994;369:321–323. doi: 10.1038/369321a0. [DOI] [PubMed] [Google Scholar]
- 27.Xue D, Horvitz H R. Nature (London) 1997;390:305–308. doi: 10.1038/36889. [DOI] [PubMed] [Google Scholar]
- 28.Boise L H, Gonzalez-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nunez G, Thompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 29.Wang K, Yin X M, Chao D T, Milliman C L, Korsmeyer S J. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 30.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Chang B S, Thompson C B, Wong S L, et al. Nature (London) 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 31.Chou J J, Li H, Salvesen G S, Yuan J, Wagner G. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 32.McDonnell J M, Fushman D, Milliman C L, Korsmeyer S J, Cowburn D. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 33.Hedgecock E M, Sulston J E, Thomson J N. Science. 1983;220:1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- 34.Conradt B, Horvitz H R. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- 35.Gu T, Orita S, Han M. Mol Cell Biol. 1998;18:4556–4564. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bloom L, Horvitz H R. Proc Natl Acad Sci USA. 1997;94:3414–3419. doi: 10.1073/pnas.94.7.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]