Abstract
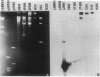
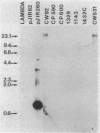
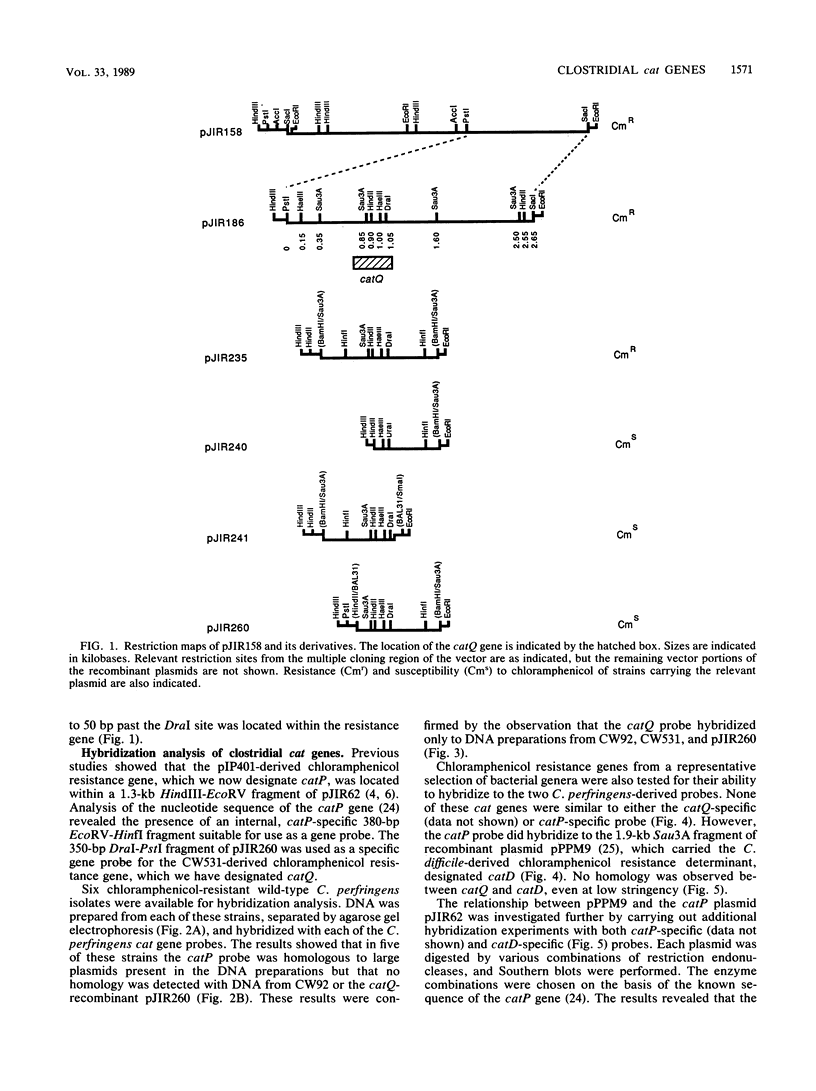
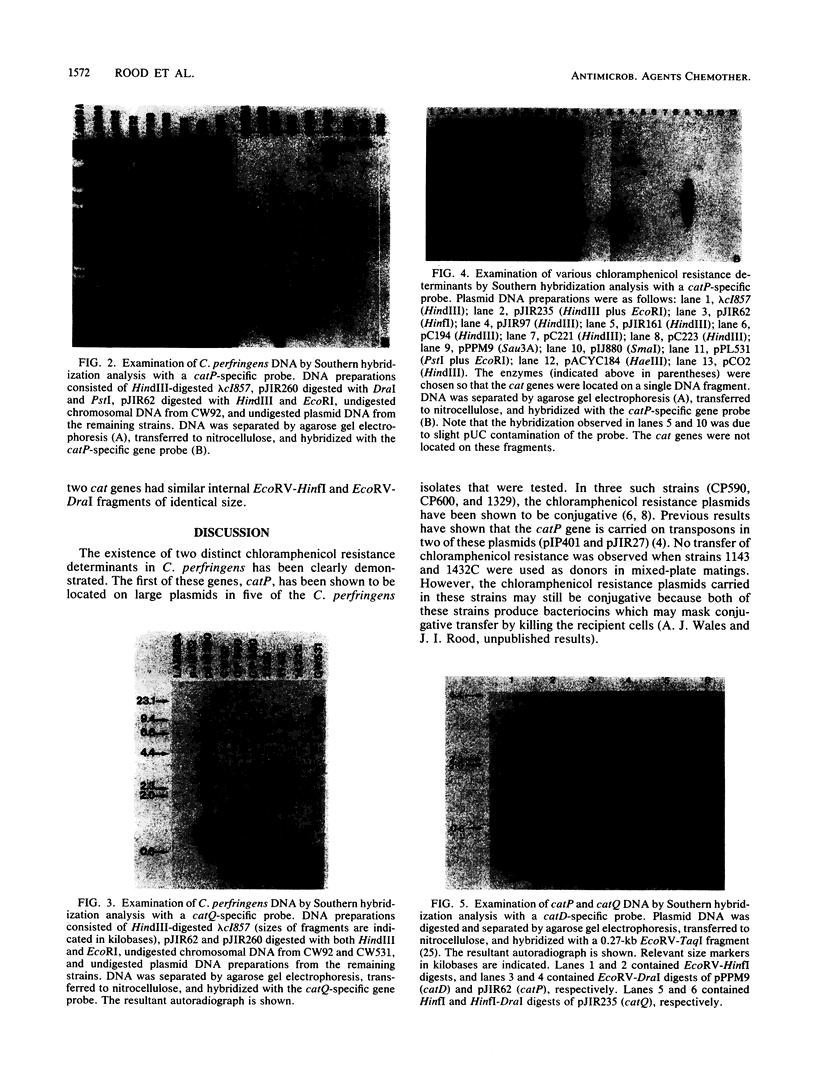
The chloramphenicol resistance determinant from a nonconjugative strain of Clostridium perfringens was cloned and shown to be expressed in Escherichia coli. Subcloning and deletion analysis localized the resistance gene, catQ, to within a 1.25-kilobase (kb) partial Sau3A fragment. The catQ gene contained internal HindII, HaeIII, and DraI restriction sites and was distinct from the catP gene, which was originally cloned (L. J. Abraham, A. J. Wales, and J. I. Rood Plasmid 14:37-46, 1985) from the conjugative C. perfringens R plasmid, pIP401. Hybridization studies were carried out with a 0.35-kb DraI-P fragment of pJIR260 as an internal catQ-specific probe and a 0.38-kb EcoRV-HinfI fragment of pJIR62 as an internal catP-specific gene probe. The results showed that the catP and catQ genes were not similar and that neither probe hybridized with cat genes from other bacterial genera. However, the catP gene was similar to the cloned catD gene from Clostridium difficile. Comparative studies with both catP and catD probes showed that these genes had significant restriction identity. We therefore suggest that these genes were derived from a common source.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham L. J., Berryman D. I., Rood J. I. Hybridization analysis of the class P tetracycline resistance determinant from the Clostridium perfringens R-plasmid, pCW3. Plasmid. 1988 Mar;19(2):113–120. doi: 10.1016/0147-619x(88)90050-9. [DOI] [PubMed] [Google Scholar]
- Abraham L. J., Rood J. I. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985 May;13(3):155–162. doi: 10.1016/0147-619x(85)90038-1. [DOI] [PubMed] [Google Scholar]
- Abraham L. J., Rood J. I. Identification of Tn4451 and Tn4452, chloramphenicol resistance transposons from Clostridium perfringens. J Bacteriol. 1987 Apr;169(4):1579–1584. doi: 10.1128/jb.169.4.1579-1584.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham L. J., Rood J. I. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J Bacteriol. 1985 Feb;161(2):636–640. doi: 10.1128/jb.161.2.636-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham L. J., Rood J. I. The Clostridium perfringens chloramphenicol resistance transposon Tn4451 excises precisely in Escherichia coli. Plasmid. 1988 Mar;19(2):164–168. doi: 10.1016/0147-619x(88)90055-8. [DOI] [PubMed] [Google Scholar]
- Abraham L. J., Wales A. J., Rood J. I. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985 Jul;14(1):37–46. doi: 10.1016/0147-619x(85)90030-7. [DOI] [PubMed] [Google Scholar]
- Berryman D. I., Rood J. I. Cloning and hybridization analysis of ermP, a macrolide-lincosamide-streptogramin B resistance determinant from Clostridium perfringens. Antimicrob Agents Chemother. 1989 Aug;33(8):1346–1353. doi: 10.1128/aac.33.8.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort G., Magot M., Ionesco H., Sebald M. Characterization and transferability of Clostridium perfringens plasmids. Plasmid. 1977 Nov;1(1):52–66. doi: 10.1016/0147-619x(77)90008-7. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J. A., Kieser H. M., Hopwood D. A. Cloning of a chloramphenicol acetyltransferase gene of Streptomyces acrimycini and its expression in Streptomyces and Escherichia coli. Gene. 1985;38(1-3):1–8. doi: 10.1016/0378-1119(85)90197-0. [DOI] [PubMed] [Google Scholar]
- Ionesco H. Transfert de la résistance à la tétracycline chez Clostridium difficile. Ann Microbiol (Paris) 1980 Mar-Apr;131A(2):171–179. [PubMed] [Google Scholar]
- Iordanescu S., Surdeanu M., Della Latta P., Novick R. Incompatibility and molecular relationships between small Staphylococcal plasmids carrying the same resistance marker. Plasmid. 1978 Sep;1(4):468–479. doi: 10.1016/0147-619x(78)90005-7. [DOI] [PubMed] [Google Scholar]
- Lyon B. R., May J. W., Skurray R. A. Analysis of plasmids in nosocomial strains of multiple-antibiotic-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1983 Jun;23(6):817–826. doi: 10.1128/aac.23.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütticken R. The genetics of GBS. Antibiot Chemother (1971) 1985;35:71–82. [PubMed] [Google Scholar]
- Rood J. I., Buddle J. R., Wales A. J., Sidhu R. The occurrence of antibiotic resistance in Clostridium perfringens from pigs. Aust Vet J. 1985 Aug;62(8):276–279. doi: 10.1111/j.1751-0813.1985.tb14251.x. [DOI] [PubMed] [Google Scholar]
- Rood J. I., Laird A. J., Williams J. W. Cloning of the Escherichia coli K-12 dihydrofolate reductase gene following mu-mediated transposition. Gene. 1980 Feb;8(3):255–265. doi: 10.1016/0378-1119(80)90003-7. [DOI] [PubMed] [Google Scholar]
- Rood J. I., Maher E. A., Somers E. B., Campos E., Duncan C. L. Isolation and characterization of multiply antibiotic-resistant Clostridum perfringens strains from porcine feces. Antimicrob Agents Chemother. 1978 May;13(5):871–880. doi: 10.1128/aac.13.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood J. I., Scott V. N., Duncan C. L. Identification of a transferable tetracycline resistance plasmid (pCW3) from Clostridium perfringens. Plasmid. 1978 Sep;1(4):563–570. doi: 10.1016/0147-619x(78)90013-6. [DOI] [PubMed] [Google Scholar]
- SMITH H. W. The effect of the continuous administration of diets containing tetracyclines and penicillin on the number of drug-resistant and drug-sensitive Clostridium welchii in the faeces of pigs and chickens. J Pathol Bacteriol. 1959 Jan;77(1):79–93. [PubMed] [Google Scholar]
- Sebald M., Bouanchaud D., Bieth G., Prévot A. R. Nature plasmidique de la résistance à plusieurs antibiotiques chez C. perfringens type A, souche 659. C R Acad Sci Hebd Seances Acad Sci D. 1975 May 26;280(20):2401–2404. [PubMed] [Google Scholar]
- Steffen C., Matzura H. Nucleotide sequence analysis and expression studies of a chloramphenicol-acetyltransferase-coding gene from Clostridium perfringens. Gene. 1989 Feb 20;75(2):349–354. doi: 10.1016/0378-1119(89)90282-5. [DOI] [PubMed] [Google Scholar]
- Wren B. W., Mullany P., Clayton C., Tabaqchali S. Molecular cloning and genetic analysis of a chloramphenicol acetyltransferase determinant from Clostridium difficile. Antimicrob Agents Chemother. 1988 Aug;32(8):1213–1217. doi: 10.1128/aac.32.8.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaidenzaig Y., Fitton J. E., Packman L. C., Shaw W. V. Characterization and comparison of chloramphenicol acetyltransferase variants. Eur J Biochem. 1979 Oct 15;100(2):609–618. doi: 10.1111/j.1432-1033.1979.tb04208.x. [DOI] [PubMed] [Google Scholar]