Abstract
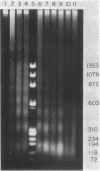
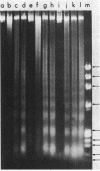
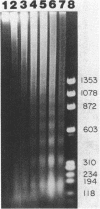
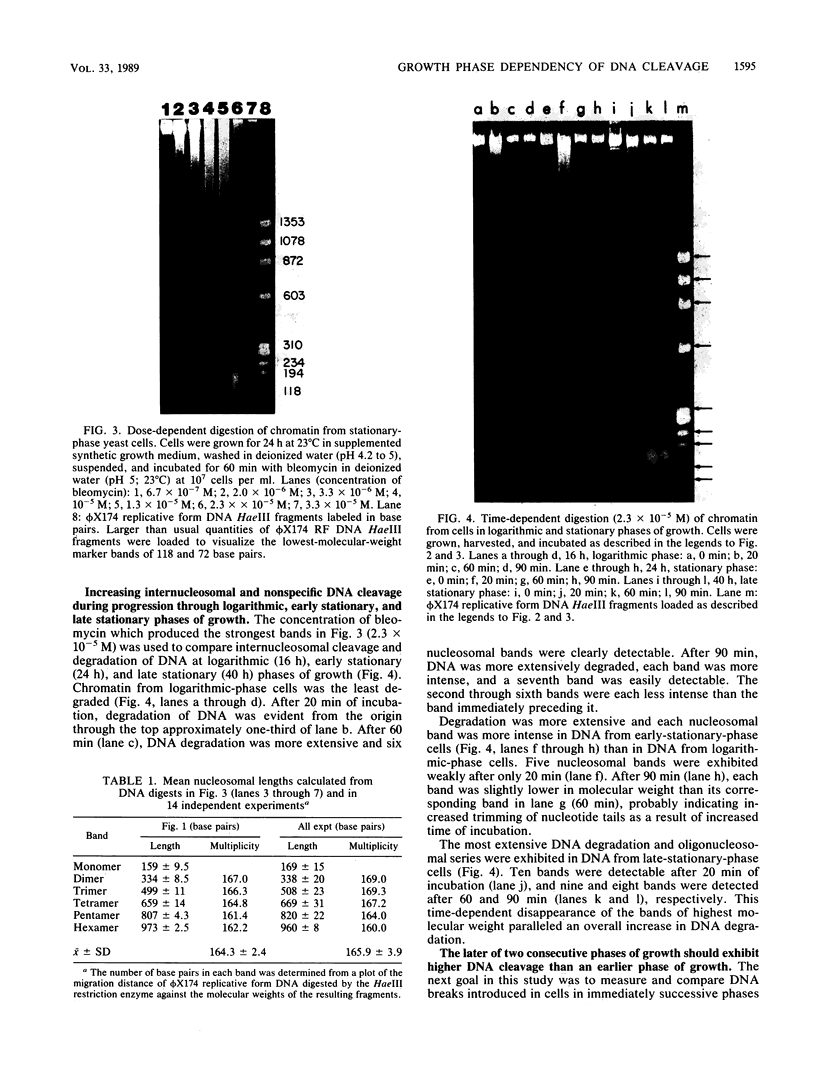
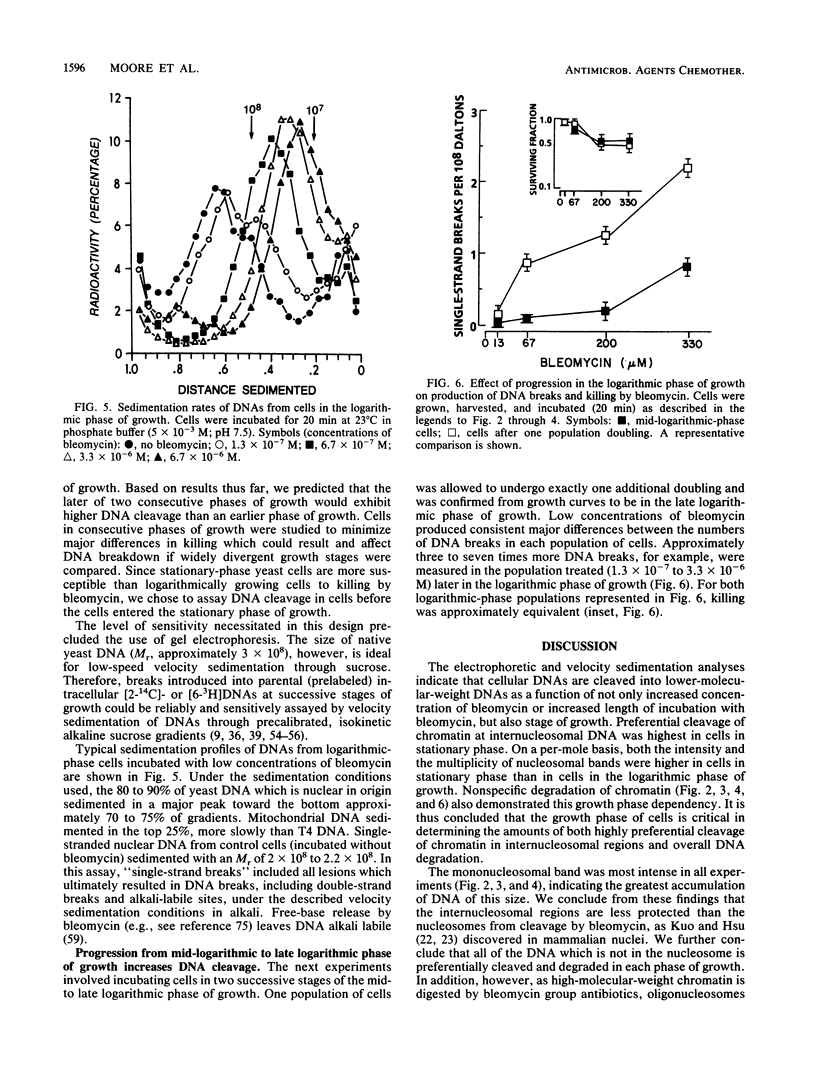
Preferential cleavage of Saccharomyces cerevisiae chromosomes in internucleosomal (linker) regions and nonspecific degradation of chromatin by an anticancer antibiotic which degrades DNA were investigated and found to increase in consecutive stages of growth. Cleavage of DNA in internucleosomal regions and intensities and multiplicities of nucleosomal bands were dependent on drug concentration, growth phase of the cells, and length of incubation. Cellular DNA was least degraded during logarithmic phase. After cells progressed only one generation in logarithmic phase, low concentrations (6.7 x 10(-7) to 3.4 x 10(-6) M) of bleomycin produced approximately three to seven times more DNA breaks. Internucleosomal cleavage was highest, and the most extended oligonucleosomal series and extensive chromatin degradation were observed during stationary phase. It is concluded that the growth phase of cells is critical in determining amounts of the highly preferential cleavage in internucleosomal regions and overall breakage and degradation of DNA. Mononucleosomal bands were most intense, indicating the greatest accumulation of DNA of this size. Mean mononucleosomal lengths were 165.9 +/- 3.9 base pairs, in agreement with yeast mononucleosomal lengths. As high-molecular-weight chromatin was digested by bleomycin, oligonucleosomes and, eventually, mononucleosomes became digested. Therefore, it is also concluded that bleomycin degradation of oligonucleosomes and trimming of DNA linker regions proceed to degradation of the monosomes (core plus linker DNA).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barranco S. C., Novak J. K., Humphrey R. M. Response of mammalian cells following treatment with bleomycin and 1,3-bis(2-chloroethyl)-1-nitrosourea during plateau phase. Cancer Res. 1973 Apr;33(4):691–694. [PubMed] [Google Scholar]
- Berry D. E., Chang L. H., Hecht S. M. DNA damage and growth inhibition in cultured human cells by bleomycin congeners. Biochemistry. 1985 Jun 18;24(13):3207–3214. doi: 10.1021/bi00334a020. [DOI] [PubMed] [Google Scholar]
- Berry D. E., Kilkuskie R. E., Hecht S. M. DNA damage induced by bleomycin in the presence of dibucaine is not predictive of cell growth inhibition. Biochemistry. 1985 Jun 18;24(13):3214–3219. doi: 10.1021/bi00334a021. [DOI] [PubMed] [Google Scholar]
- Burger R. M., Berkowitz A. R., Peisach J., Horwitz S. B. Origin of malondialdehyde from DNA degraded by Fe(II) x bleomycin. J Biol Chem. 1980 Dec 25;255(24):11832–11838. [PubMed] [Google Scholar]
- Burger R. M., Peisach J., Horwitz S. B. Mechanism of bleomycin action: in vitro studies. Life Sci. 1981 Feb 16;28(7):715–727. doi: 10.1016/0024-3205(81)90153-3. [DOI] [PubMed] [Google Scholar]
- Carrier W. L., Setlow R. B. Paper strip method for assaying gradient fractions containing radioactive macromolecules. Anal Biochem. 1971 Oct;43(2):427–432. doi: 10.1016/0003-2697(71)90272-7. [DOI] [PubMed] [Google Scholar]
- Drewinko B., Patchen M., Yang L. Y., Barlogie B. Differential killing efficacy of twenty antitumor drugs on proliferating and nonproliferating human tumor cells. Cancer Res. 1981 Jun;41(6):2328–2333. [PubMed] [Google Scholar]
- Ehmann U. K., Lett J. T. Review and evaluation of molecular weight calculations from the sedimentation profiles of irradiated DNA. Radiat Res. 1973 Apr;54(1):152–162. [PubMed] [Google Scholar]
- Forte M. A., Fangman W. L. Naturally occurring cross-links in yeast chromosomal DNA. Cell. 1976 Jul;8(3):425–431. doi: 10.1016/0092-8674(76)90155-0. [DOI] [PubMed] [Google Scholar]
- Forte M. A., Fangman W. L. Naturally occurring cross-links in yeast chromosomal DNA. Cell. 1976 Jul;8(3):425–431. doi: 10.1016/0092-8674(76)90155-0. [DOI] [PubMed] [Google Scholar]
- Franco L., Johns E. W., Navlet J. M. Histones from baker's yeast. Isolation and fractionation. Eur J Biochem. 1974 Jun 1;45(1):83–89. doi: 10.1111/j.1432-1033.1974.tb03532.x. [DOI] [PubMed] [Google Scholar]
- Giloni L., Takeshita M., Johnson F., Iden C., Grollman A. P. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J Biol Chem. 1981 Aug 25;256(16):8608–8615. [PubMed] [Google Scholar]
- Grollman A. P., Takeshita M. Interactions of bleomycin with DNA. Adv Enzyme Regul. 1980;18:67–83. doi: 10.1016/0065-2571(80)90009-6. [DOI] [PubMed] [Google Scholar]
- Haidle C. W., Weiss K. K., Kuo M. T. Release of free bases from deoxyribonucleic acid after reaction with bleomycin. Mol Pharmacol. 1972 Sep;8(5):531–537. [PubMed] [Google Scholar]
- Keller T. J., Oppenheimer N. J. Enhanced bleomycin-mediated damage of DNA opposite charged nicks. A model for bleomycin-directed double strand scission of DNA. J Biol Chem. 1987 Nov 5;262(31):15144–15150. [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kuo M. T. Altered gel electrophoretic mobility of bleomycin-mediated release of nucleosomal DNA. Cancer Res. 1981 Jun;41(6):2433–2438. [PubMed] [Google Scholar]
- Kuo M. T., Hsu T. C. Bleomycin causes release of nucleosomes from chromatin and chromosomes. Nature. 1978 Jan 5;271(5640):83–84. doi: 10.1038/271083a0. [DOI] [PubMed] [Google Scholar]
- Kuo M. T. Preferential damage of active chromatin by bleomycin. Cancer Res. 1981 Jun;41(6):2439–2443. [PubMed] [Google Scholar]
- Lohr D. E. Detailed analysis of the nucleosomal organization of transcribed DNA in yeast chromatin. Biochemistry. 1981 Oct 13;20(21):5966–5972. doi: 10.1021/bi00524a007. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Yeast chromatin subunit structure. Science. 1975 Apr 11;188(4184):165–166. doi: 10.1126/science.1090006. [DOI] [PubMed] [Google Scholar]
- Mauro F., Falpo B., Briganti G., Elli R., Zupi G. Effects of antineoplastic drugs on plateau-phase cultures of mammalian cells. II. Bleomycin and hydroxyurea. J Natl Cancer Inst. 1974 Mar;52(3):715–722. doi: 10.1093/jnci/52.3.715. [DOI] [PubMed] [Google Scholar]
- Mirabelli C. K., Huang C. H., Fenwick R. G., Crooke S. T. Quantitative measurement of single- and double-strand breakage of DNA in Escherichia coli by the antitumor antibiotics bleomycin and talisomycin. Antimicrob Agents Chemother. 1985 Apr;27(4):460–467. doi: 10.1128/aac.27.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabelli C. K., Huang C. H., Prestayko A. W., Crooke S. T. Structure-activity relationships involved in the site-specific fragmentation of linear duplex DNAs by talisomycin and bleomycin analogs. Cancer Chemother Pharmacol. 1982;8(1):57–65. doi: 10.1007/BF00292872. [DOI] [PubMed] [Google Scholar]
- Mirabelli C. K., Ting A., Huang C. H., Mong S., Crooke S. T. Bleomycin and talisomycin sequence-specific strand scission of DNA: a mechanism of double-strand cleavage. Cancer Res. 1982 Jul;42(7):2779–2785. [PubMed] [Google Scholar]
- Miyamoto T., Terasima T. Effects of bleomycin on Burkitt lymphoma cells grown in vitro and in vivo. Jpn J Cancer Res. 1986 Jul;77(7):674–681. [PubMed] [Google Scholar]
- Moore C. W. Bleomycin-induced DNA repair by Saccharomyces cerevisiae ATP-dependent polydeoxyribonucleotide ligase. J Bacteriol. 1988 Oct;170(10):4991–4994. doi: 10.1128/jb.170.10.4991-4994.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W. Control of in vivo (cellular) phleomycin sensitivity by nuclear genotype, growth phase, and metal ions. Cancer Res. 1982 Mar;42(3):929–933. [PubMed] [Google Scholar]
- Moore C. W. Internucleosomal cleavage and chromosomal degradation by bleomycin and phleomycin in yeast. Cancer Res. 1988 Dec 1;48(23):6837–6843. [PubMed] [Google Scholar]
- Moore C. W. Ligase-deficient yeast cells exhibit defective DNA rejoining and enhanced gamma ray sensitivity. J Bacteriol. 1982 Jun;150(3):1227–1233. doi: 10.1128/jb.150.3.1227-1233.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W., Little J. B. Rapid and slow DNA rejoining in nondividing human diploid fibroblasts treated with bleomycin and ionizing radiation. Cancer Res. 1985 May;45(5):1982–1986. [PubMed] [Google Scholar]
- Moore C. W., Malcolm A. W., Tomkinson K. N., Little J. B. Ultrarapid recovery from lethal effects of bleomycin and gamma-radiation in stationary-phase human diploid fibroblasts. Cancer Res. 1985 May;45(5):1978–1981. [PubMed] [Google Scholar]
- Moore C. W. Modulation of bleomycin cytotoxicity. Antimicrob Agents Chemother. 1982 Apr;21(4):595–600. doi: 10.1128/aac.21.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W. Responses of radiation-sensitive mutants of Saccharomyces cerevisiae to lethal effects of bleomycin. Mutat Res. 1978 Aug;51(2):165–180. doi: 10.1016/s0027-5107(78)80016-5. [DOI] [PubMed] [Google Scholar]
- Moore C. W., Schmick A. Genetic effects of impure and pure saccharin in yeast. Science. 1979 Sep 7;205(4410):1007–1010. doi: 10.1126/science.382356. [DOI] [PubMed] [Google Scholar]
- Moore C. W., Vossler D. A. Influence of pH and length of post-treatment incubation on bleomycin-induced DNA damage. Biochim Biophys Acta. 1980 Dec 11;610(2):425–429. doi: 10.1016/0005-2787(80)90024-6. [DOI] [PubMed] [Google Scholar]
- Moore C. W. cdc9 ligase-defective mutants of Saccharomyces cerevisiae exhibit lowered resistance to lethal effects of bleomycin. J Bacteriol. 1982 Sep;151(3):1617–1620. doi: 10.1128/jb.151.3.1617-1620.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris N. R. A comparison of the structure of chicken erythrocyte and chicken liver chromatin. Cell. 1976 Dec;9(4 Pt 1):627–632. doi: 10.1016/0092-8674(76)90045-3. [DOI] [PubMed] [Google Scholar]
- Murray V., Martin R. F. The sequence specificity of bleomycin-induced DNA damage in intact cells. J Biol Chem. 1985 Sep 5;260(19):10389–10391. [PubMed] [Google Scholar]
- Murugesan N., Xu C., Ehrenfeld G. M., Sugiyama H., Kilkuskie R. E., Rodriguez L. O., Chang L. H., Hecht S. M. Analysis of products formed during bleomycin-mediated DNA degradation. Biochemistry. 1985 Oct 8;24(21):5735–5744. doi: 10.1021/bi00342a008. [DOI] [PubMed] [Google Scholar]
- Nelson D. A., Beltz W. R., Rill R. L. Chromatin subunits from baker's yeast: isolation and partial characterization. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1343–1347. doi: 10.1073/pnas.74.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. A., Oosterhof D. K., Rill R. L. Histones H2A and H2B are neighbors along DNA in chromatin: characterization of subnucleosomal particles containing H2A+H2B. Nucleic Acids Res. 1977 Dec;4(12):4223–4234. doi: 10.1093/nar/4.12.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R. G., Fangman W. L. Nucleosome organization of the yeast 2-micrometer DNA plasmid: a eukaryotic minichromosome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6515–6519. doi: 10.1073/pnas.76.12.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Saunders C., Davie J. R., Hamkalo B. A. Ultrastructural organization of yeast chromatin. J Cell Biol. 1982 Apr;93(1):217–222. doi: 10.1083/jcb.93.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray G. R., Hahn G. M., Bagshaw M. A., Kurkjian S. Cell survival and repair of plateau-phase cultures after chemotherapy--relevance to tumor therapy and to the in vitro screening of new agents. Cancer Chemother Rep. 1973 Nov-Dec;57(4):473–475. [PubMed] [Google Scholar]
- Resnick M. A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976 Jan 16;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Reynolds R. J., Friedberg E. C. Molecular mechanism of pyrimidine dimer excision in Saccharomyces cerevisiae. I. Studies with intact cells and cell-free systems. Basic Life Sci. 1980;15:121–139. doi: 10.1007/978-1-4684-3842-0_8. [DOI] [PubMed] [Google Scholar]
- Reynolds R. J., Friedberg E. C. Molecular mechanisms of pyrimidine dimer excision in Saccharomyces cerevisiae: incision of ultraviolet-irradiated deoxyribonucleic acid in vivo. J Bacteriol. 1981 May;146(2):692–704. doi: 10.1128/jb.146.2.692-704.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. D., Seale R. L., Yu J. Transcribed chromatin exhibits an altered nucleosomal spacing. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5505–5509. doi: 10.1073/pnas.80.18.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Nagai K., Yamaki H., Tanaka N., Umezawa H. On the mechanism of action of bleomycin: scission of DNA strands in vitro and in vivo. J Antibiot (Tokyo) 1969 Sep;22(9):446–448. doi: 10.7164/antibiotics.22.446. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C., Isenberg I. The organization of oligonucleosomes in yeast. Nucleic Acids Res. 1983 Jun 11;11(11):3717–3736. doi: 10.1093/nar/11.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takita T., Muraoka Y., Fujii A., Ito H., Maeda K. The structure of the sulfur-containing chromophore of phleomycin, and chemical transformation of phleomycin to bleomycin. J Antibiot (Tokyo) 1972 Mar;25(3):197–199. doi: 10.7164/antibiotics.25.197. [DOI] [PubMed] [Google Scholar]
- Takita T., Muraoka Y., Nakatani T., Fujii A., Umezawa Y., Naganawa H. Chemistry of bleomycin. XIX Revised structures of bleomycin and phleomycin. J Antibiot (Tokyo) 1978 Aug;31(8):801–804. doi: 10.7164/antibiotics.31.801. [DOI] [PubMed] [Google Scholar]
- Takita T., Muraoka Y., Yoshioka T., Fujii A., Maeda K. The chemistry of bleomycin. IX. The structures of bleomycin and phleomycin. J Antibiot (Tokyo) 1972 Dec;25(12):755–757. doi: 10.7164/antibiotics.25.755. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Tien Kuo M., Haidle C. W., Inners L. D. Characterization of bleomycin-resistant DNA. Biophys J. 1973 Dec;13(12):1296–1306. doi: 10.1016/s0006-3495(73)86063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien Kuo M., Hsu T. C. Biochemical and cytological studies of bleomycin actions on chromatin and chromosomes. Chromosoma. 1978 Sep 1;68(3):229–240. doi: 10.1007/BF00335418. [DOI] [PubMed] [Google Scholar]
- Tobüren D. Differential survival of cell subpopulations in spheroids after treatment with bleomycin. J Cancer Res Clin Oncol. 1981;102(2):189–193. doi: 10.1007/BF00410671. [DOI] [PubMed] [Google Scholar]
- Twentyman P. R., Bleehen N. M. The sensitivity of cells in exponential and stationary phases of growth to bleomycin and to 1,3-bis(2-chloroethyl)-1-nitrosourea. Br J Cancer. 1973 Dec;28(6):500–507. doi: 10.1038/bjc.1973.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twentyman P. R. Bleomycin--mode of action with particular reference to the cell cycle. Pharmacol Ther. 1983;23(3):417–441. doi: 10.1016/0163-7258(83)90022-0. [DOI] [PubMed] [Google Scholar]
- Twentyman P. R. Comparative chemosensitivity of exponential- versus plateau-phase cells in both in vitro model systems. Cancer Treat Rep. 1976 Dec;60(12):1719–1722. [PubMed] [Google Scholar]
- Wu J. C., Kozarich J. W., Stubbe J. The mechanism of free base formation from DNA by bleomycin. A proposal based on site specific tritium release from Poly(dA.dU). J Biol Chem. 1983 Apr 25;258(8):4694–4697. [PubMed] [Google Scholar]