Abstract
Alternate splicing, leading to the insertion of the tripeptide KTS in the linker between the third and fourth C2H2 zinc fingers, changes both the DNA-binding function and the subnuclear localization of the Wilms' tumor suppressor protein (WT1). We have used NMR relaxation experiments to determine the molecular basis for the differing DNA recognition properties of the WT1–KTS and WT1+KTS isoforms. Our results show that the KTS insertion increases the flexibility of the linker between fingers 3 and 4 and abrogates binding of the fourth zinc finger to its cognate site in the DNA major groove. This represents a mechanism whereby a single zinc-finger gene can be used, through alternate splicing, to fulfill different functions in the cell.
The Wilms' tumor suppressor gene encodes a DNA-binding protein containing four Cys2His2 zinc fingers (1, 2) and is essential for normal mammalian urogenital development (3). Mutations or deletions of this gene are associated with Wilms' tumor, one of the most common pediatric solid tumors (4, 5), and frequently are associated with congenital defects of the genitourinary tract such as Denys-Drash syndrome. Alternate splicing, leading to the insertion of the tripeptide KTS in the linker between the third and fourth C2H2 zinc fingers (6), changes both the DNA-binding function and the subnuclear localization of the Wilms' tumor suppressor protein (WT1) (7–10).
Four isoforms of WT1 are formed by alternate RNA splicing (6). In one splice variant, an additional 17 aa are inserted N-terminal to the first zinc finger but these have no known effect on protein function. The other alternate splicing event results in the insertion (+KTS isoform) or omission (−KTS isoform) of the KTS tripeptide in the canonical TGEKP linker sequence between zinc fingers 3 and 4 (Fig. 1). All four isoforms are present in cells expressing WT1 and their relative abundance is constant; indeed, developmental abnormalities have been observed in patients with altered ratios of +KTS and −KTS isoforms (4, 11). Insertion of the KTS tripeptide has a profound effect on both the DNA-binding affinity and the specificity of WT1. The WT1–KTS isoform binds to a 9-bp early growth response protein (EGR-1) consensus site with high affinity whereas the +KTS splice variant binds to the same site 10- to 20-fold more weakly (7, 8). The different WT1 isoforms localize to distinct compartments in the nucleus; WT1–KTS colocalizes with other transcription factors, whereas the more abundant +KTS isoform associates with components of the pre-mRNA-splicing machinery (small nuclear ribonucleoproteins) where it potentially functions through RNA binding (9). Thus, the presence or absence of the KTS insert in the third linker modulates both the DNA-binding affinity and the functional distribution of WT1 within the nucleus. Indeed, it has been suggested that differences in DNA-binding affinity of the +KTS and −KTS splice variants might dictate the pattern of nuclear localization, with the tighter-binding −KTS isoform being preferentially compartmentalized with the DNA (10).
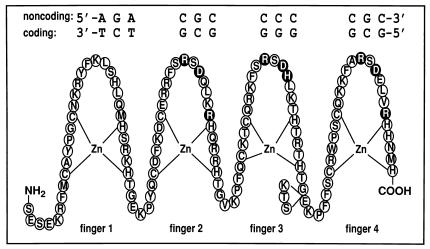
Figure 1.
Schematic representation of the WT1 zinc-finger domain (fingers 1–4; residues 318–438) with the core DNA recognition element showing the “antiparallel” orientation in which the complex is formed (the 5′ end of the coding strand of the DNA is aligned with the C terminus of the WT1 protein). The circles representing the putative DNA base-contacting residues (14, 23, 35) are shaded black. Base-contacting residues in finger 1 are unknown, and those in finger 4 occur only in the −KTS isoform (see text). The KTS insertion site between fingers 3 and 4 is indicated.
In this report, we examine the structural basis for the difference in DNA recognition and binding properties that dictate the respective in vivo regulatory roles of the two KTS isoforms of WT1. Understanding of the molecular mechanism by which alternate splicing can modulate the biological function of a zinc-finger transcription factor is clearly of major importance. To this end, we have used NMR relaxation measurements to examine the interactions of both the +KTS and −KTS isoforms of WT1 with its cognate DNA. Of the four WT1 zinc fingers, fingers 2–4 are closely similar in sequence to the three zinc fingers encoded by the early growth response gene, EGR-1 and bind to the EGR-1 recognition element (GCG GGG GCG). Heteronuclear NMR spectra were recorded for WT1+KTS and WT1–KTS, both free and bound to a 14-bp oligonucleotide duplex containing an EGR-1-binding site. The DNA sequence (Fig. 1) was selected to give high binding affinity for the +KTS isoform, based on the observed sequence preferences for interaction with finger 1 (12, 13).
Materials and Methods
Preparation of Protein and DNA.
Two uniformly 15N-labeled protein isoforms that encompass the zinc-finger domain (residues 318–438, plus an N-terminal Ala) of WT1, which included (+KTS) or omitted (−KTS) residues 408–410, were expressed and purified as described (14). Complexes were formed with a 14-bp DNA duplex (coding strand sequence 5′-CGCGGGGGCGTCTG-3′) prepared from complementary synthetic oligonucleotides (Operon Technologies, Alameda, CA; Keystone Biosource, Camarillo, CA). A detailed protocol for this procedure has been described (14). All NMR samples were prepared in argon-saturated d11-Tris buffer (10 mM, pH 6.7) in a mixture of 95% H2O/5% 2H2O, containing 20 mM KCl, 50 μM ZnSO4, 2 mM NaN3, 0.2 mM DSS. The sample concentrations for the free proteins were 0.8 mM, and the WT1–DNA complexes were at equimolar concentrations of 0.3 mM.
NMR Measurements.
The [15N] longitudinal relaxation time (T1), 15N transverse relaxation time (T2), and [1H]-[15N] nuclear Overhauser effect (NOE) were measured by using standard inversion-recovery, Carr-Purcell-Meiboom-Gill, and steady-state NOE experiments (15) with water flipback (16). Spectra were recorded at 310 K on a Bruker DRX spectrometer operating at a 1H frequency of 600.13 MHz and equipped with an 8-mm probe (Nalorec Cryogenics, Martinez, CA). For T1, spectra were acquired at 13 relaxation delay times for the DNA complexes (30 ms to 3 s), and 15 delay times for the free proteins (30 ms to 2.5 s); for T2, 17 delay times were acquired for both complexes (6–482 ms) and the free proteins (6–642 ms). Duplicate data sets were recorded at three time points for both T1 and T2 to allow estimation of errors. The refocusing delay in the Carr-Purcell-Meiboom-Gill experiment was 1 ms. The [1H]-[15N] NOE experiment was repeated five times for each complex and three times for the free proteins, and data sets with and without 1H saturation were interleaved. Recovery delays of 3.5, 3.5, and 2.5 s were used for the T1, T2, and NOE experiments, respectively. All spectra were processed by using felix (Molecular Simulations, San Diego, CA), and all T1 and T2 time constants were calculated from the decay curves by using the program curvefit (A.G. Palmer, III, Columbia University, New York). Published backbone resonance assignments were used (14), although those of the free proteins were extrapolated from 293 to 310 K by using a series of [15N] heteronuclear single quantum coherence spectra recorded at intervening temperatures.
Analysis of Relaxation Results.
Reduced spectral densities were calculated by using published methods, assuming a 1/ω2 dependence at high frequencies (17). An effective rotational correlation time, τm, for each residue was calculated from the spectral densities (18). A mean correlation time for each zinc finger was calculated by averaging individual τm values for residues in elements of regular secondary structure (β-hairpin, α-helix).
Results and Discussion
The dynamics of the WT1 polypeptide chain were investigated by measurement of the [15N] T1, [15N] T2, and the [1H]-[15N] heteronuclear NOE by using two-dimensional inverse-detected NMR experiments. For each [15N]–[1H] pair, the data provide information on the fast internal dynamics as well as the overall tumbling of the macromolecule. The heteronuclear [1H]-[15N] NOE of the backbone amides, shown as a function of residue number in Fig. 2, is highly sensitive to motions of the polypeptide backbone on a ps to ns time scale. NOE values smaller than ≈0.6 indicate large amplitude backbone fluctuations on a ps to ns time scale. For the free protein, pronounced minima are observed in the heteronuclear NOE values at each linker (Fig. 2), showing that the linkers are all highly flexible. Insertion of the KTS tripeptide in the linker between fingers 3 and 4 significantly increases the backbone flexibility.
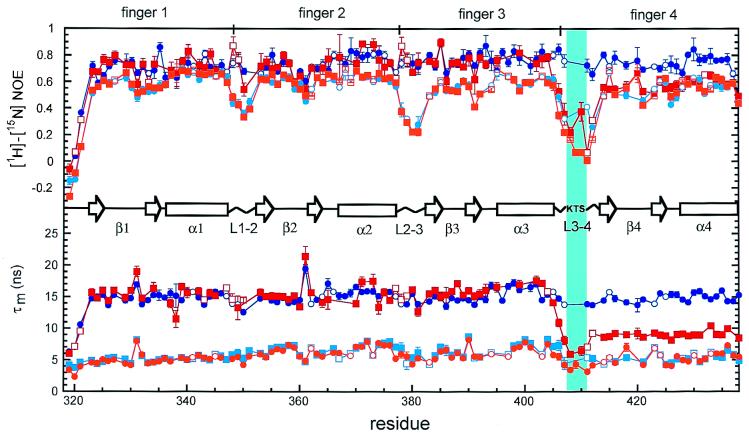
Figure 2.
Backbone [1H]-[15N] NOE and effective τm values for free (light colors) and DNA-bound (dark colors) +KTS (squares) and −KTS (circles) as a function of residue number. The β-sheet (open arrows), α-helix (open rectangles), linker regions (zigzag lines), and alternative KTS splice site (vertical cyan bar) in the zinc-finger domain of WT1 are indicated schematically. Error bars on the [1H]-[15N] NOE data points represent the SEM in the respective measurement, whereas the error bars on the τm plot account for uncertainties in both the measured data points and the respective exponential fits used to obtain T1 and T2 for each residue (67% confidence). Open symbols indicate resonances that are overlapped and for which there are consequently large uncertainties in the relaxation parameters.
On binding to DNA, the average NOE increases for all four zinc fingers of WT1–KTS because of loss of the domain flexibility that is a characteristic property of free zinc-finger proteins (19, 20). Furthermore, the short time scale motions in all three linkers are damped and the heteronuclear NOEs in these regions become comparable in magnitude to those in the finger domains (Fig. 2). Damping of linker motions on DNA binding also has been observed for other zinc-finger proteins and is caused by DNA-induced helix capping interactions and formation of a highly conserved structure by the linker residues (20, 21). This DNA-induced folding of the linker has been likened to a “snap lock” that, once activated, helps to restrict the motions between adjacent fingers and position them for optimal binding interactions in the major groove (21).
Very different behavior is observed for the WT1+KTS isoform. On DNA binding, increases are observed in the heteronuclear NOE for residues in fingers 1–3 and in the linkers between fingers 1 and 2 and between fingers 2 and 3; these changes are very similar to those seen on forming the WT1–KTS complex with DNA and indicate similar restriction of backbone motions on DNA binding. However, the linker containing the KTS insertion, between fingers 3 and 4, retains the high degree of backbone flexibility seen in the free WT1+KTS isoform and the NOEs in finger 4 closely parallel those of the free protein.
The most revealing insights into the effect of alternate splicing on the DNA-binding properties come from a consideration of the τm calculated from the relaxation data. These data are shown for both isoforms in Fig. 2. Not surprisingly, the τm is considerably smaller in both WT1 isoforms free in solution (6.0 ± 1.1 ns and 5.8 ± 1.0 ns for the WT1–KTS and WT1+KTS isoforms, respectively) compared with the same proteins bound to the DNA recognition element. The average τm for zinc fingers 1–4 in the DNA complex of WT1–KTS is 15 ± 1.0 ns, reflecting the higher molecular mass (≈25 kDa, vs. ≈15 kDa for the free protein) and slower molecular tumbling of the complex. For the WT1+KTS complex, the average τm for zinc fingers 1–3 (15 ± 1.4 ns) is the same as for the −KTS isoform, confirming that these three fingers are tightly bound to the DNA. Remarkably, finger 4 tumbles in solution with an effective τm that is very much shorter (τm = 9.2 ± 0.6 ns) than the τm for fingers 1–3. Indeed, this τm value is much closer to that of the free protein than that of the protein–DNA complex, providing unequivocal evidence that binding of WT1+KTS to DNA does not significantly restrict the molecular tumbling of finger 4. This can only occur if finger 4 makes very weak interactions or no interactions with the DNA. Thus, the [15N] relaxation data show clearly that the effect of the KTS insert in the linker between fingers 3 and 4 is to prevent finger 4 from making optimal interactions with its cognate binding site in the DNA major groove. The short τm value associated with finger 4 indicates that, at best, it makes only transient interactions with DNA and tumbles largely independently of the finger 1–3–DNA complex, as shown schematically in Fig. 3.
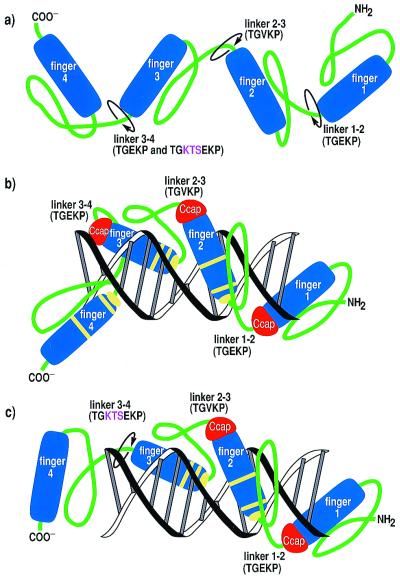
Figure 3.
Cartoon drawing illustrating the structural and dynamic consequences of the KTS insertion on DNA binding by WT1. (a) Free WT1 isoforms and DNA-bound (b) −KTS and (c) +KTS proteins have different degrees of interdomain tumbling motions. Circular arrows in a and c illustrate the sites of linker flexibility between all four fingers in the free proteins (a) and in linker 3–4 of the +KTS complex with DNA (c). Yellow bands in b and c indicate putative base-contacting residues. Note that finger 4 does not contact the DNA in the +KTS complex (c).
Thus, the molecular basis for the decreased DNA-binding affinity of the WT1+KTS isoform lies in the increased length and flexibility of the linker between fingers 3 and 4, which prevents finger 4 from binding to its cognate site on the DNA, even though its structure and DNA contact residues are unchanged by the splicing event (14). With the exception of potential phosphate backbone contacts involving the Lys side chain (22), the residues in the highly conserved TGEKP linker are distant from the DNA in the available structures of zinc-finger protein complexes (22–26). Loss of finger 4 binding is unlikely to arise solely from disruption of the Lys-phosphate backbone contacts, because mutagenesis studies of transcription factor IIIA suggest that these contribute relatively little to the overall DNA-binding affinity (27). A more likely explanation is that insertion of the KTS tripeptide prevents the linker from adopting the unique and highly conserved structure observed for all TGEKP linkers in the available x-ray and NMR structures of zinc finger–DNA complexes (21). In addition, the increased entropic cost of ordering the longer linker would militate against localization and binding of finger 4 in the DNA major groove.
It might be expected that the longer linker in the WT1+KTS isoform could alter the DNA-binding specificity to favor a target with increased spacing between the finger 3- and 4-binding sites. This appears not to be the case, however; screening of complete genomic DNA libraries failed to identify efficient binding targets for WT1+KTS (8). For all known recognition elements, the −KTS isoform binds DNA with higher affinity than the +KTS isoform (7, 8, 12). Thus, abrogation of finger 4–DNA interactions by the KTS insert is likely to be universal to all DNA sequences that WT1 binds. We note that construction of a molecular model suggests that the inserted linker residues could easily be accommodated structurally without displacement of either of the neighboring fingers from their DNA-binding sites. This provides added support for our contention that changes in linker conformation and increased linker flexibility play a major role in mediating the DNA-binding properties of WT1+KTS.
Our data thus provide insights into the molecular mechanism by which the DNA-binding properties, subnuclear localization, and function of the Wilms' tumor gene product are mediated by alternate splicing. Whereas WT1–KTS binds EGR-like recognition elements with high affinity and is clearly involved in transcriptional regulation, the +KTS isoform binds DNA weakly and is preferentially associated with the splicing machinery where, it is postulated, it may interact with RNA (9). Although RNA sequences have been identified that bind with high affinity to WT1+KTS (28), a physiologically relevant target sequence has yet to be identified. It is also possible that WT1+KTS may function by binding protein components of the splicing machinery. Indeed, it has recently been shown that WT1 interacts with the splicing factor U2AF65 in vitro, and that the +KTS isoform binds more strongly than WT1–KTS (29). Further characterization of the detailed molecular function and interactions of WT1+KTS is of particular interest because variations in the ratio of ± KTS splice isoforms are associated with human disease. Frasier Syndrome is caused by a mutation in an intronic region of the WT1 gene that prevents production of the +KTS isoform (30, 31). Although many questions still remain about the physiological interactions mediated by the different isoforms, the studies presented here reveal the fundamental molecular mechanism by which alternate splicing modulates DNA binding and hence determines the function and cellular localization of WT1.
Finally, we note that multiplicity of function is not without precedent in zinc-finger proteins. Transcription factor IIIA, with nine zinc fingers, binds both the internal control region of the 5S RNA gene and the 5S RNA transcript itself (32, 33). In this case, different sets of zinc fingers (fingers 1–3 and fingers 4–7, respectively) mediate the high-affinity interactions with DNA and RNA (34). Alternate splicing to alter the length of a linker represents a further simple but very effective mechanism by which the DNA-binding properties and hence the biological function of a zinc-finger protein can be modulated.
Acknowledgments
We thank J. Chung for assistance with NMR measurements and B. Duggan for computer programs used in the reduced spectral mapping and many helpful discussions. This work was supported by Grant GM36643 from the National Institutes of Health. J.H.L. is supported by Fellowship CA79192 from the National Institutes of Health.
Abbreviations
- WT1
Wilms' tumor suppressor protein
- EGR-1
early growth response protein
- NOE
nuclear Overhauser effect
- T1
longitudinal relaxation time
- T2
transverse relaxation time
- τm
effective rotational correlation time
- DSS
2,2-dimethyl-2-silapentane-5-sulfonate, sodium salt
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gessler M, Poustka A, Cavenee W, Neve R L, Orkin S H, Bruns G A P. Nature (London) 1990;343:774–778. doi: 10.1038/343774a0. [DOI] [PubMed] [Google Scholar]
- 2.Call K M, Glaser T, Ito C Y, Buckler A J, Pelletier J, Haber D A, Rose E A, Kral A, Yeger H, Lewis W H, et al. Cell. 1990;60:509–520. doi: 10.1016/0092-8674(90)90601-a. [DOI] [PubMed] [Google Scholar]
- 3.Kreidberg J A, Sariola H, Loring J M, Maeda M, Pelletier J, Housman D, Jaenisch R. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 4.Coppes M J, Campbell C E, Williams B R G. FASEB J. 1993;7:886–894. doi: 10.1096/fasebj.7.10.8393819. [DOI] [PubMed] [Google Scholar]
- 5.Hastie N D. Annu Rev Genet. 1994;28:523–558. doi: 10.1146/annurev.ge.28.120194.002515. [DOI] [PubMed] [Google Scholar]
- 6.Haber D A, Sohn R L, Buckler A J, Pelletier J, Call K M, Housman D E. Proc Natl Acad Sci USA. 1991;88:9618–9622. doi: 10.1073/pnas.88.21.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rauscher F J, Morris J F, Tournay O E, Cook D M, Curran T. Science. 1990;250:1259–1262. doi: 10.1126/science.2244209. [DOI] [PubMed] [Google Scholar]
- 8.Bickmore W A, Oghene K, Little M H, Seawright A, van Heyningen V, Hastie N D. Science. 1992;257:235–237. doi: 10.1126/science.1321494. [DOI] [PubMed] [Google Scholar]
- 9.Larsson S H, Charlieu J-P, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie N D. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- 10.Charlieu J-P, Larsson S, Miyagawa K, van Heyningen V, Hastie N D. J Cell Sci. 1995;19:95–99. doi: 10.1242/jcs.1995.supplement_19.14. [DOI] [PubMed] [Google Scholar]
- 11.Reddy J C, Licht J D. Biochim Biophys Acta. 1996;1287:1–28. doi: 10.1016/0304-419x(95)00014-7. [DOI] [PubMed] [Google Scholar]
- 12.Drummond I A, Rupprecht H D, Rohwer-Nutter P, Lopez-Guisa J M, Madden S L, Rauscher F J, III, Sukhatme V P. Mol Cell Biol. 1994;14:3800–3809. doi: 10.1128/mcb.14.6.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton T B, Borel F, Romaniuk P J. Biochemistry. 1998;37:2051–2058. doi: 10.1021/bi9717993. [DOI] [PubMed] [Google Scholar]
- 14.Laity J H, Dyson H J, Wright P E. Biochemistry. 2000;39:5341–5348. doi: 10.1021/bi9926678. [DOI] [PubMed] [Google Scholar]
- 15.Farrow N A, Zhang O, Forman-Kay J D, Kay L E. Biochemistry. 1995;34:868–878. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- 16.Grzesiek S, Bax A. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 17.Farrow N A, Zhang O, Szabo A, Torchia D A, Kay L E. J Biomol NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 18.Bracken C, Carr P A, Cavanagh J, Palmer A G., III J Mol Biol. 1999;285:2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 19.Brüschweiler R, Liao X, Wright P E. Science. 1995;268:886–889. doi: 10.1126/science.7754375. [DOI] [PubMed] [Google Scholar]
- 20.Foster M P, Wuttke D S, Radhakrishnan I, Case D A, Gottesfeld J M, Wright P E. Nat Struct Biol. 1997;4:605–608. doi: 10.1038/nsb0897-605. [DOI] [PubMed] [Google Scholar]
- 21.Laity J H, Dyson H J, Wright P E. J Mol Biol. 2000;295:719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- 22.Wuttke D S, Foster M P, Case D A, Gottesfeld J M, Wright P E. J Mol Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 23.Pavletich N P, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 24.Pavletich N P, Pabo C O. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 25.Houbaviy H B, Usheva A, Shenk T, Burley S K. Proc Natl Acad Sci USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim C A, Berg J M. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 27.Choo Y, Klug A. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caricasole A, Duarte A, Larsson S H, Hastie N D, Little M, Holmes G, Todorov I, Ward A. Proc Natl Acad Sci USA. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies R C, Calvio C, Bratt E, Larsson S H, Lamond A I, Hastie N D. Genes Dev. 1998;12:3217–3225. doi: 10.1101/gad.12.20.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbaux S, Niaudet P, Gubler M C, Grunfeld J P, Jaubert F, Kuttenn F, Fekete C N, Souleyreau-Therville N, Thibaud E, Fellous M, et al. Nat Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- 31.Klamt B, Koziell A, Poulat F, Wieacker P, Scambler P, Berta P, Gessler M. Hum Mol Genet. 1998;7:709–714. doi: 10.1093/hmg/7.4.709. [DOI] [PubMed] [Google Scholar]
- 32.Honda B M, Roeder R G. Cell. 1980;22:119–126. doi: 10.1016/0092-8674(80)90160-9. [DOI] [PubMed] [Google Scholar]
- 33.Pelham H R, Brown D D. Proc Natl Acad Sci USA. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clemens K R, Wolf V, McBryant S J, Zhang P, Liao X, Wright P E, Gottesfeld J M. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 35.Elrod-Erickson M, Rould M A, Nekludova L, Pabo C O. Structure (London) 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]