Abstract
Ser-15 of human p53 (corresponding to Ser-18 of mouse p53) is phosphorylated by ataxia-telangiectasia mutated (ATM) family kinases in response to ionizing radiation (IR) and UV light. To determine the effects of phosphorylation of endogenous murine p53 at Ser-18 on biological responses to DNA damage, we introduced a missense mutation (Ser-18 to Ala) by homologous recombination into both alleles of the endogenous p53 gene in mouse embryonic stem (ES) cells. Our analyses showed that phosphorylation of murine p53 at Ser-18 in response to IR or UV radiation was required for a full p53-mediated response to these DNA damage-inducing agents. In contrast, phosphorylation of p53 at Ser-18 was not required for ATM-dependent cellular resistance after exposure to IR. Additionally, efficient acetylation of the C terminus of p53 in response to DNA damage did not require phosphorylation of murine p53 at Ser-18.
Activation of the tumor suppressor protein p53 as a transcription factor in response to DNA damage is postulated to be a critical event in preventing cancer. In response to DNA damage or other cellular stresses, p53 protein levels and its activity as a transcription factor increase. The stabilization and activation of p53 results in the arrest of cell-cycle progression in late G1 or in apoptosis in a manner that depends on cell type and the severity of cellular damage; however, our understanding of the mechanisms by which these responses occur is incomplete (1, 2). Recent studies show that DNA damage-activated signaling pathways lead to the phosphorylation of p53 at both N- and C-terminal sites, and accumulating evidence suggests that p53 phosphorylation plays an important role in regulating both the stability and activity of p53 (3).
One event that is postulated to be critical for p53-mediated DNA damage responses is phosphorylation of serine 15 (Ser-15) in human p53 (4, 5). Ser-15 is an evolutionarily conserved residue, corresponding to Ser-18 in murine p53, that can be phosphorylated in vitro by several related protein kinases belonging to the ataxia-telangiectasia mutated (ATM) family (8); these include DNA-PK (6), ATR (7), and ATM itself (8–11). ATM is the product of a tumor suppressor gene that is mutated in the autosomally recessive human genetic disease ataxia-telangiectasia (A-T). A-T patients exhibit pleiotropic defects, including hypersensitivity to ionizing radiation (IR) and a high incidence of cancer (8). Cells derived from A-T patients, as well as those from ATM-deficient mice, exhibit delayed or reduced responses to IR, including defects in both S-phase and G2/M cell-cycle checkpoints (12, 13). Recent studies have shown that the rapid phosphorylation of human p53 at Ser-15 after IR-induced DNA damage requires the ATM protein kinase (9–11). Furthermore, cells lacking the ATM kinase are hypersensitive to IR (8). In contrast, ATM-deficient cells respond normally to DNA damage caused by exposure to UV light and to other physical and physiological stresses; however, Ser-15 may be phosphorylated by another ATM family member, ATR, in response to UV treatment (7). These findings suggest that Ser-15 phosphorylation may be a master regulator of p53's responses to DNA damage.
Although the delayed and reduced phosphorylation of human p53 at Ser-15 in ATM-deficient cells exposed to IR is correlated with a reduced accumulation of p53 and suggests that phosphorylation of Ser-15 is responsible, at least in part, for p53 stabilization (9–11), the roles of Ser-15 phosphorylation in p53-mediated responses to DNA damage have remained controversial. Although one study reported that phosphorylation of p53 at Ser-15 disrupted its interactions with Mdm2, which would stabilize p53 (4), others have presented contrary evidence and argue instead that Ser-15 phosphorylation is required for the acetylation of p53 at C-terminal Lys residues (14, 15). Acetylation is believed to activate the sequence-specific DNA binding ability of p53, a step that is necessary for most, if not all, p53 responses. Yet others argue that phosphorylation of Ser-20 is critical for inhibiting Mdm2-mediated degradation of p53 (16, 17). Ser-20 of human p53 recently was shown to be phosphorylated by Chk1 and Chk2 (18, 19), which are activated by ATM and required for p53 stabilization after IR (19–21). Therefore, phosphorylation of p53 at Ser-20 through this indirect ATM-dependent pathway also may contribute to ATM-dependent p53 stabilization.
To address the importance of Ser-15 phosphorylation for these processes in a physiological context, we used genetic approaches to introduce a missense mutation into the endogenous p53 gene of mouse embryonic stem (ES) cells that changes Ser-18 of mouse p53 to alanine, a residue that cannot be phosphorylated. Analysis of the mutant murine ES cells before and after differentiation indicated that phosphorylation of mouse p53 at Ser-18 is required for a full p53-mediated response induced by exposure to either IR or UV light. Thus, our findings support a role for Ser-18 phosphorylation in p53 stabilization after DNA damage; however, we also show that this phosphorylation is not required for ATM-dependent cellular sensitivity to IR nor for acetylation at the two C-terminal sites after DNA damage.
Materials and Methods
Construction of the Targeting Vector.
Ser-18 of p53 is encoded within exon 2 of the mouse p53 gene (ref. 22; and Fig. 1A). A mouse p53 genomic DNA fragment containing exon 2 was isolated, and the nucleotides encoding Ser-18 were changed to encode Ala by site-directed mutagenesis using a kit (Stratagene). The mutated p53 genomic segment was used to construct a targeting vector by inserting the phosphoglycerol kinase (PGK)-neomycin resistance gene (PGK-neor gene) flanked by two LoxP sites into a unique SalI site in intron 4, and the thymidine kinase gene was inserted at one end of the p53 genomic DNA to allow negative selection of randomly integrated inserts. An EcoRI site also was introduced into intron 1 by site-directed mutagenesis as an aid to determine that the wild-type exon had been replaced (Fig. 1B).
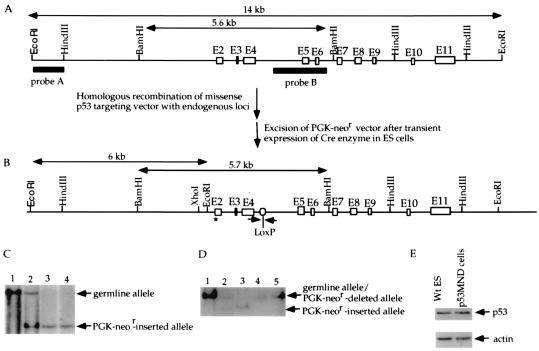
Figure 1.
Introduction of a missense mutation into the endogenous p53 gene of ES cells to change Ser-18 to Ala. (A) The mouse germ-line p53 locus. Blank boxes represent the p53 exons and two filled bars represent the probes for Southern blot analysis of the mutant/deleted allele. The germ-line 14-kb EcoRI fragment and 5.6-kb BamHI fragment are indicated. (B) Mutant p53 allele with the PGK-neor segment deleted. The size of the PGK-neor-deleted mutant BamHI fragment and the positions of PCR primers (arrows) flanking the remaining LoxP site are indicated. (C) Southern blotting analysis of the genomic DNA derived from wild-type (lane 1), heterozygous mutant (with the PGK-neor segment inserted; lane 2), and homozygous mutant (with the PGK-neor segment inserted; lanes 3 and 4) ES cells. Genomic DNA was digested with EcoRI and hybridized with probe A. The positions of restriction fragments from the germ-line and mutant alleles are indicated with arrowheads. (D) Southern blotting analysis of genomic DNA derived from wild-type (lane 1), heterozygous mutant (with the PGK-neor segment inserted; lane 2), homozygous mutant (with the PGK-neor segment inserted; lane 3), and p53MND ES cells (lanes 4 and 5). Genomic DNA was digested with BamHI and hybridized with probe B. The positions of the BamHI fragments derived from the wild-type/PGK-neor-deleted mutant allele and the PGK-neor-inserted mutant allele are indicated. (E) Western blotting analysis of the constitutive p53 protein level in wild-type and p53MND ES cells. To ensure that equal amounts of protein were loaded, the blot was stripped and probed with anti-actin antibody. Lanes are labeled on the top; the primary antisera used (p53 or actin) is indicated at the right.
Generation of Heterozygous, Homozygous, and p53MND Mutant ES Cells.
The targeting construct was linearized with XbaI and electroporated into 20 × 106 J-1 ES cells as described (13). Transfectants were selected with G418 (0.3 mg/ml) and gancyclovir (1 μM). Homologous recombination events were confirmed by Southern blotting with probe A after EcoRI digestion, which gives a 14-kb germ-line band and a 6.5-kb mutant band (Fig. 1 A and B). To generate homozygous mutant ES cells, heterozygous mutant ES cells were cultured under the selection of increasing concentrations of G418 as described (13). ES cell colonies surviving 4.8 mg/ml G418 were expanded and screened for homozygous mutants by the Southern blot method. To delete the PGK-neor gene from both alleles of the homozygous mutant ES cells, 20 μg of a circular plasmid that drives expression of the Cre enzyme was transiently transfected into the PGK-neor-inserted homozygous mutant ES cells as described (23). Transfectants were plated at a density of 500–1,000 colonies per 6-cm plate, and ES cell colonies were screened for the Cre-mediated deletion by PCR with primers indicated in Fig. 1B. Positive ES cells identified by PCR were subcloned and subsequently confirmed by Southern blotting after digestion with BamHI and hybridization to probe B, which revealed a 5.6-kbp germ-line band, a 4.8-kbp PGK-neor-inserted band, or a 5.7-kbp PGK-neor-deleted band.
Culture and Treatment of ES Cells and Differentiated ES Cells.
Before irradiation treatment, ES cells were cultured without a feeder layer in DMEM supplemented with 15% FCS, glutamine, nonessential amino acids, antibiotics, 100 μM β-mercaptoethanol, and recombinant lymphocyte inhibitory factor (LIF). ES cells were irradiated with 5, 10, or 20 Gy γ-ray or exposed to 30 or 60 J/M2 UV radiation and harvested at various times after treatment. Differentiation of ES cells in vitro with retinoic acid was performed as described (24). Briefly, subconfluent ES cell culture were trypsinized, and individual ES cells were plated onto gelatinized 10-cm plates at a density of 2 million cells per plate in ES cell culture medium supplemented with 3 × 10−7 M retinoic acid but without LIF and a feeder layer. Most cells in the culture were differentiated after 4–5 days of treatment with retinoic acid.
Western Blot Analysis.
Protein extract from 4 × 105 cells per sample was separated by 12% SDS/PAGE and transferred to a nitrocellulose membrane. The membrane was blocked with 5% dry milk and probed with a monoclonal antibody against p53 (Pab240; Santa Cruz Biotechnology) or a polyclonal antibody against p21 (Santa Cruz Biotechnology). The filter subsequently was incubated with horseradish peroxide-conjugated secondary antibody, developed with enhanced chemiluminescence PLUS (ECL PLUS), scanned with a Storm system (Molecular Dynamics), and quantitated by using the imagequant program (Molecular Dynamics). The filter subsequently was exposed to x-ray film. To standardize the amount of protein in each lane, the filter was stripped and probed with a polyclonal antibody against actin (Santa Cruz Biotechnology), developed with ECL Plus, and quantitated as described above.
Cell Cycle Analysis.
Differentiated ES cells were irradiated with 20 Gy γ-irradiation. The p53-dependent cell-cycle G1 arrest after irradiation was analyzed by staining with propidium iodide (Sigma) for DNA content and with anti-BrdUrd antibody (Southern Biotechnology Associates) for DNA synthesis as described (13).
Clonogenic Survival and Proliferation Assay.
Wild-type, p53MND, ATM−/−, and p53−/− ES cells were plated on mouse embryonic fibroblast feeder layers in 6-well plates at a density of 1 × 103 cells/well. Twenty-four hours after plating, ES cells were exposed to different doses of γ-irradiation. The irradiated cells were cultured with a change of medium every 2 days. Colonies were counted after 8 days of culture, and the ratio of the number of surviving colonies in the irradiated sample to those in the untreated control was calculated. Proliferative assays were performed essentially as described (13). Briefly, wild-type, p53MND, and p53−/− differentiated ES cells were plated in 35-mm plates at a density of 1 × 105 cells/plate. Three plates were trypsinized and counted each day afterward.
Phosphorylation- and Acetylation-Specific Mouse p53 Antibodies.
PAbSer(P)15 rabbit polyclonal antibody specific for mouse p53 phosphorylated at Ser-18 (human Ser-15) has been described (25, 26). PAbLys(Ac)317 M and PAbLys(Ac)379 M antibodies specific for mouse p53 acetylated at Lys-317 or Lys-379 were prepared similarly. Rabbits were immunized with the mouse p53 acetylated peptide Ac-310–322(317Ac)C [i.e., Ac-Ser-Ala-Ser-Pro-Pro-Gln-Lys-Lys(Ac)-Lys-Pro-Leu-Asp-Gly-Cys-NH2] or Ac-374–386(379Ac)C [i.e., Ac-Thr-Ser-Arg-His-Lys-Lys(Ac)-Thr-Met-Val-Lys-Lys-Val-Gly-Cys-NH2], and acetylation site-specific antibodies were affinity purified by use of the corresponding SulfoLinked acetylated peptides. The specificity of each antibody was confirmed by ELISA and immunoblot assays. The detection of acetylated p53 was performed as described (26).
Results
Construction of ES Cells with a Germ-Line Missense Mutation That Changes Ser-18 to Ala.
To introduce a missense mutation into exon 2 of the endogenous p53 gene of mouse ES cells that changed Ser-18 to alanine, nucleotides AGC encoding Ser-18 were changed to GCC encoding alanine. The mutated genomic DNA fragment then was used to construct a targeting vector (Fig. 1 A and B). Homologous recombination between the endogenous p53 locus and the targeting vector led to the replacement of the germ-line exon 2 with the mutant exon 2 harboring the missense mutation (Ser-18 to Ala), and this event was confirmed by Southern blot analysis (Fig. 1C). Homozygous mutant (both alleles mutated) ES cells were generated by culturing the heterozygous mutant ES cells in increasing concentrations of G418 as described (21), and the result was confirmed by Southern blotting (Fig. 1C). The PGK-neor gene in the mutant p53 alleles can suppress expression of the mutant p53 allele (data not shown); therefore the PGK-neor gene was excised from the genome through transient expression of the Cre enzyme (23) (Fig. 1 B and D). Deletion of the PGK-neor gene from both alleles was confirmed by Southern blot analysis (Fig. 1 A, B, and D); we refer to these as p53 mutant-neor-deleted (p53MND) ES cells. That these cells expressed only p53 with Ala in place of Ser-18 was confirmed by sequence analysis of amplified exon segments and the p53 cDNA. In addition, analysis of p53 protein levels in p53MND and wild-type ES cells indicated that the missense mutation did not affect the basal expression level or stability of the p53 protein (Fig. 1E).
Phosphorylation of p53 at Ser-18 in ES Cells After DNA Damage.
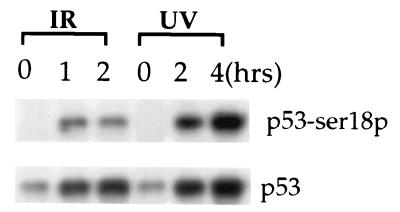
Ser-15 of human p53 was shown to be phosphorylated in human cell lines after UV and IR treatment (4, 9, 10). Therefore, we tested whether murine p53 becomes phosphorylated at Ser-18, the equivalent site to Ser-15 in human p53, after damage induced by UV and IR treatment. Using an antibody specific for murine p53 phosphorylated at Ser-18, our results indicated that Ser-18 becomes phosphorylated in murine p53 in response to DNA damage, which is analogous to the behavior of human p53 (Fig. 2).
Figure 2.
Phosphorylation of mouse p53 at Ser-18 in ES cells at various times after IR or UV treatment. ES cells were exposed to 5 Gy IR or 60 J/M2 UV light. Cell extracts from irradiated cells and untreated controls were analyzed by Western blotting with antibodies specific for p53 or p53 phosphorylated at Ser-18. The phosphorylated p53 (Top) and total p53 are indicated. Time after irradiation is indicated at the top of each lane.
Accumulation of p53 and p21 After DNA Damage Is Defective in p53MND Cells.
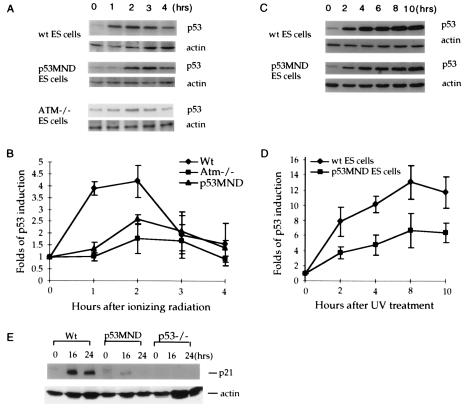
Similar to other cell types such as mouse embryonic fibroblasts, wild-type ES cells showed rapid, but transient, induction of p53 after IR with a 4- to 5-fold increase in protein levels 1 h after treatment (13, 27) (Fig. 3 A and B). p53 levels in mock-treated ES cells did not increase (data not shown). The p53 induction in response to IR in ES cells also depends on ATM because there was a delayed and greatly reduced accumulation of p53 in response to IR in ATM−/− ES cells (13) (Fig. 3 A and B). Analysis of the kinetics and extent of p53 induction in p53MND ES cells indicated that there was essentially no induction of the p53 protein levels 1 h post-IR (Fig. 3 A and B). In addition, the maximum accumulation of p53 in the p53MND ES cells after IR was only about half that seen in the wild-type ES cells (Fig. 3 A and B). A similar reduction in accumulation also was seen after exposure to 10 or 20 Gy (data not shown), indicating that the reduction is independent of dose. These findings indicate that p53 accumulation in p53MND ES cells after IR is delayed and reduced as seen in the ATM−/− ES cells.
Figure 3.
p53 and p21 expression in p53MND cells after IR (A and B) or UV (C and D). (A and C) Protein levels of p53 at different times after irradiation. No difference was seen in the constitutive basal level of p53 in the untreated ES cells at various time points. Genotypes of ES cells are indicated on the left and primary antibody (p53 or actin) is indicated on the right. (B and D) Quantitative analysis of p53 induction at the indicated times after IR or UV treatment in ES cells. The blot was developed with ECL Plus, scanned, and quantitated as described in Materials and Methods. Mean values with standard deviations from three independent experiments are plotted. (E) p21 expression in wild-type, p53MND, and p53−/− differentiated ES cells. Times after treatment, genotypes, and the primary antibody used (p21 or actin) are given.
Ser-18 of p53 also is phosphorylated by ATM-family kinases after UV treatment (7). Because ES cells show typical p53 accumulation after DNA damage induced by UV treatment (27, 28), we also analyzed the kinetics and extent of p53 induction in wild-type and p53MND ES cells after UV treatment. Similar to p53 responses to IR in p53MND ES cells, the increase in p53 protein after UV treatment was attenuated, and the maximum increase was only 50–60% of that seen in wild-type ES cells (Fig. 3 C and D). Consistent with the above results, p53 induction in differentiated ES cells also was reduced after UV treatment (data not shown). Therefore, these findings support the notion that phosphorylation of p53 at Ser-18 is required for efficient p53 accumulation after UV treatment.
Differentiated ES cells undergo typical p53-dependent p21 induction after DNA damage (27). To test the effects of the Ser-18 to Ala change on p53-dependent G1 cell-cycle arrest, we differentiated wild-type and p53MND ES cells in culture as described (24, 27) and examined their ability to induce p21 after UV treatment. As expected, p21 induction in differentiated ES cells after DNA damage depended on p53 because there was no p21 induction in p53−/− differentiated ES cells post-UV radiation (Fig. 3E). The level of p21 increased significantly in wild-type cells at 16 and 24 h after UV irradiation, whereas induction of p21 at 16 h after UV radiation in the p53MND cells was attenuated significantly compared with that in wild-type cells (Fig. 3E). In addition, there was little induction of p21 in p53MND cells at 24 h after UV radiation (Fig. 3E). These results are consistent with the findings that activation of p53 is impaired in the p53MND mutant cells after UV treatment (Fig. 3E).
Role of Ser-18 Phosphorylation in G1 Arrest, ATM-Dependent Hypersensitivity to IR and Cellular Proliferation.
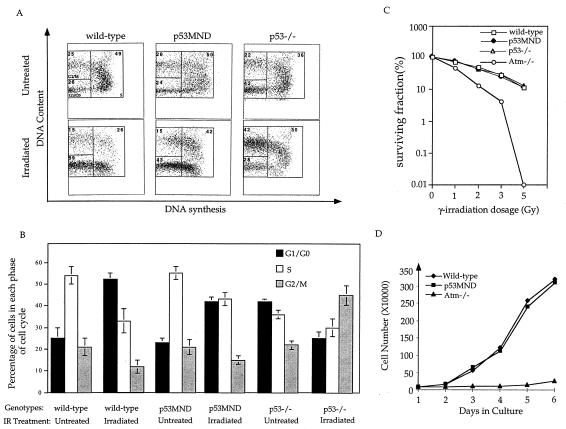
Differentiated ES cells undergo p53-dependent cell-cycle G1 arrest after DNA damage. To test the effects of the Ser-18 to Ala change on p53-mediated G1 arrest, we differentiated wild-type, p53−/−, and p53MND ES cells in culture and examined their ability to undergo G1/S cell-cycle arrest after IR as described (13). Because synchronizing cells at G1/G0 through serum starvation might introduce additional cellular stresses that activate signaling pathways affecting p53 stabilization, the differentiated ES cells were asynchronously growing at the time of irradiation treatment. In wild-type differentiated ES cells, there was a 35% decrease in S-phase cells whereas G1/G0-phase cells more than doubled at 12 h after IR treatment, indicating that the differentiated wild-type ES cells arrest their cell cycle at the G1/S border after DNA damage (Fig. 4 A and B). This cell-cycle G1 arrest depends on p53 because there was no G1 arrest in the p53−/− differentiated ES cells after the same irradiation treatment (Fig. 4 A and B). In differentiated p53MND ES cells, S-phase cells decreased by only 15% at the same time, whereas the extent of accumulation of G1/G0-phase cells was correspondingly less than that in wild-type cells (Fig. 4 A and B). Therefore, consistent with the finding that p53 induction in p53MND ES cells is reduced after DNA damage, differentiated p53MND ES cells are impaired in their p53-dependent cell-cycle arrest after exposure to IR.
Figure 4.
Cell cycle analysis, survival, and proliferation of wild-type, p53−/−, and p53MND cells after DNA damage. (A) A representative FACS analysis of untreated or irradiated differentiated wild-type, p53−/−, and p53MND ES cells at 12 h after treatment with 20 Gy IR. (B) Summary of the percentage of cells in various cell-cycle phases in untreated or irradiated wild-type, p53MND, and p53−/− differentiated ES cells. Mean values with standard deviations from three independent experiments are shown. (C) Clonogenic survival of wild-type, p53−/−, Atm−/−, and p53MND ES cells after exposure to increasing dosage of IR. For each dosage, colonies from duplicated wells were counted. Consistent data were obtained from three independent experiments. (D) Proliferation of wild-type, Atm−/−, and p53MND differentiated ES cells. The cell number represents the average from three plates for each time point. Consistent data were obtained from two independent experiments. Genotypes are indicated.
Both human and mouse ATM-deficient cells are hypersensitive to IR (8). To test whether ATM-mediated phosphorylation of p53 at Ser-18 is required for this event, we compared the hypersensitivity of wild-type, p53−/−, p53MND, and ATM−/− ES cells to IR as described (13). Although ATM−/− ES cells are hypersensitive to IR, p53−/−, p53MND, and wild-type ES cells showed a similar sensitivity to IR (Fig. 4C).
Human and mouse ATM−/− cells are defective in their cellular proliferation partly because of the impaired cell-cycle G1/S transition (8, 13). Because p53 is involved in regulating the G1/S transition, we examined the cellular proliferation characteristics of wild-type, p53MND, and ATM−/− differentiated ES cells. Although ATM−/− differentiated ES cells proliferated very slowly, p53MND and wild-type differentiated ES cells proliferated with the same rapid kinetics (Fig. 4D). Taken together, these results indicate that phosphorylation of p53 at Ser-18 does not contribute to hypersensitivity to IR nor to ATM-dependent cell cycle progression.
Phosphorylation of p53 at Ser-18 Does Not Affect the Acetylation of p53 at Lys-317 and Lys-379.
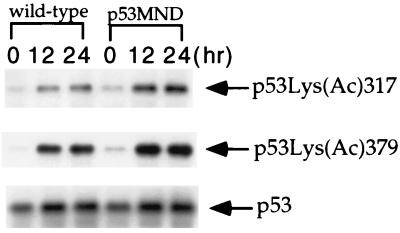
Several reports have suggested that phosphorylation of p53 at Ser-15 might activate the acetylation of human p53 at Lys-320 and Lys-382 (corresponding to mouse Lys-317 and Lys-379) through increased recruitment of transcription coactivators such as p300 and CBP (14, 15). To determine whether phosphorylation of murine p53 at Ser-18 regulates the acetylation of C-terminal sites in a similar way, we analyzed the acetylation of p53 at Lys-317 and Lys-379 in wild-type and p53MND differentiated ES cells after UV treatment. In both wild-type and p53MND cells, p53 was acetylated to a similar extent at 16 and 24 h after UV treatment, indicating that phosphorylation of p53 at Ser-18 is not essential for acetylation of p53 at the C terminus in response to DNA damage (Fig. 5).
Figure 5.
Acetylation of p53 at Lys-317 and -379 in wild-type and p53MND differentiated ES cells at 12 and 24 h after UV radiation. Differentiated ES cells were exposed to 60 J/M2 UV light and treated with 5 μM trichostatin A for 4 h before harvesting. Cell extracts from the irradiated and untreated controls were analyzed by Western blot with antibodies specific for p53 or p53 acetylated at Lys-317 or Lys-379. Acetylated p53 (Top and Middle) and total p53 (Bottom) are indicated with arrowheads.
Discussion
To determine the importance of phosphorylation of human p53 at the ATM phosphorylation site (Ser-15), we used homologous recombination and LoxP/Cre-mediated deletion to create mutant murine ES cells that express an endogenous p53 that cannot be phosphorylated at this site (Ser-18 of murine p53). This genetic approach has several advantages. First, only primary cells with defined genetic backgrounds are used, which minimizes genetic variability and the possibility that inadvertent mutants may contribute to the phenotype. Second, expression of the mutant p53 is driven by its own promoter and regulatory elements; thus, the problem of deregulated expression of p53 that is typical of transient transfections and nonhomologous, stable transformations is avoided. Using this system, we show that phosphorylation of murine p53 at Ser-18 is required to achieve an efficient and maximum p53 DNA damage response after exposure to either IR or UV light. This conclusion is supported by our finding that p53-dependent functions, including the induction of p21/Waf1 expression and G1 cell-cycle arrest, were partially defective in mutant differentiated ES cells expressing p53, which could not be phosphorylated at Ser-18. Disruption of the ability of Mdm2 to target p53 degradation is the primary cause of p53 induction after DNA damage in both undifferentiated and differentiated ES cells (29). Our findings are consistent with a contribution of phosphorylation at Ser-18 to that process; however, our results do not distinguish between a direct effect of Ser-18 phosphorylation on p53 stability and indirect effects Ser-18 phosphorylation might have by, e.g., affecting other modifications that regulate p53 stability. In addition, knock-in mice with this Ser-18A mutation are being generated to test the effects of this mutation on the tumor suppression activity of p53.
Several previous in vitro studies have examined the effects of mutating one or several phosphorylation sites within p53 on the biological activity of the protein. In one study, overexpression of a human p53 Ser-15 to Ala mutant protein was reported to result in a partial failure of the checkpoint that inhibits cell cycle progression (30). Another study showed that simultaneous mutation of Ser-9, -18, and -37 of murine p53 (corresponding to Ser-6, -15, and -33 in human p53) significantly reduced the ability of p53 to suppress transformation of rat embryo fibroblasts transfected with E1A and Ras (31). More recently, Ser-15 and Ser-20 were shown to play a role in p53-mediated apoptosis (32). Moreover, using the transactivation domain of p53 fused to a heterologous DNA-binding domain, it was shown that loss of Ser-15 resulted in a 4- to 5-fold inhibition of transactivation activity (14). In contrast to the above reports, others have shown that it is possible to mutate many p53 phosphorylation sites without significantly affecting either the induction or function of the protein (33, 34). These conflicting data most likely resulted from differences in experimental conditions and the genetic backgrounds of the cell lines used by different investigators. In particular, overexpression of p53 mutants in transient and stable transfectants could lead to negative selection of transfectants expressing high p53 levels, which makes it hard to control for the activation of unidentified signaling pathways affecting p53 induction or function.
Although our findings indicate that the phosphorylation of mouse p53 at Ser-18 by ATM family kinases is required for an efficient p53 response to IR, the observation that p53 accumulation in mouse ATM−/− cells after IR is more dramatically reduced than it is in p53MND cells suggests that ATM-mediated phosphorylation of other substrates or other p53 sites is also necessary for a full p53 response to IR-induced DNA damage. Consistent with this notion, recent findings indicate that phosphorylation of human p53 at Ser-20, which is indirectly regulated by ATM through the downstream effector Chk1/2 kinases, may at least partially control p53 stabilization and regulation of its activity (16–21). Furthermore, the ATM kinase clearly has other functional targets independent of p53 because cells deficient in ATM exhibit defects in G1/S as well as S phase and G2 phase checkpoints, and recently ATM was shown to be required for a rapid phosphorylation of Mdm2 in response to IR (35). Such targets presumably account for the findings of ourselves and others that whereas p53−/− and p53MND ES cells have the same sensitivity to IR as the wild-type cells, ATM-deficient ES cells are hypersensitive to IR and this hypersensitivity is p53-independent (8, 24, 28).
Acetylation of human p53 at Lys-320 by PCAF and Lys-373 and/or Lys-382 by p300/CBP, is thought to contribute to the activation of p53's site-specific DNA-binding activity (26, 36–38). Phosphorylation of Ser-15 has been proposed as a mechanism that permits subsequent modification of the C-terminal lysine residues through the recruitment of p300/CBP/PCAF (14, 15, 26). Therefore, the activation of acetylation in response to DNA damage was a second potential mechanism for which Ser-15 phosphorylation might be required. However, our results clearly show that p53 with the Ser-18 to Ala change is acetylated in response to UV light. Although this result does not argue against the notion that the N terminus of p53 is involved in recruitment of the histone acetylases that acetylate p53, it clearly shows that phosphorylation of Ser-18 alone is not required to promote this DNA damage induced response. Therefore, the defective induction of p21 in p53MND cells after UV treatment most likely is caused by reduced p53 accumulation rather than impaired recruitment of p300/CBP. Future studies involving the mutation of the endogenous murine p53 gene to prevent phosphorylation at other N-terminal sites will be required to address this issue.
Acknowledgments
This work was partially supported by grants from the U.S. Army Medical Research and University of California Breast Cancer Research Program (to Y.X.). C.W.A. was supported in part by National Institutes of Health Grant GM52825 at Brookhaven National Laboratory under contract with the U.S. Department of Energy.
Abbreviations
- IR
ionizing radiation
- ES
embryonic stem
- PGK
phosphoglycerol kinase
- ATM
ataxia-telangiectasia mutated
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220252297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220252297
References
- 1.Prives C, Hall P A. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Meek D W. Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- 4.Shieh S Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 5.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotman G, Shiloh Y. Cancer Surv. 1997;29:285–304. [PubMed] [Google Scholar]
- 9.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 10.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa K, Taya Y, Tamai K, Yamaizumi M. Mol Cell Biol. 1999;19:2828–2834. doi: 10.1128/mcb.19.4.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Baltimore D. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 14.Dumaz N, Meek D W. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert P F, Kashanchi F, Radonovich M F, Shiekhattar R, Brady J N. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 16.Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shieh S Y, Taya Y, Prives C. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shieh S Y, Ahn J, Tamai K, Taya Y, Prives C. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 19.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 20.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 21.Matsuoka S, Huang M, Elledge S J. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 22.Bienz B, Zakut-Houri R, Givol D, Oren M. EMBO J. 1984;3:2179–2183. doi: 10.1002/j.1460-2075.1984.tb02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Davidson L, Alt F W, Baltimore D. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 24.Aladjem M I, Spike B T, Rodewald L W, Hope T J, Klemm M, Jaenisch R, Wahl G M. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 25.Jimenez G S, Bryntesson F, Torres-Arzayus M I, Priestley A, Beeche M, Saito S, Sakaguchi K, Appella E, Jeggo P A, Taccioli G E, et al. Nature (London) 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]
- 26.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabapathy K, Klemm M, Jaenisch R, Wagner E F. EMBO J. 1997;16:6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corbet S W, Clarke A R, Gledhill S, Wyllie A H. Oncogene. 1999;18:1537–1544. doi: 10.1038/sj.onc.1202436. [DOI] [PubMed] [Google Scholar]
- 29.Chao C, Saito S, Kang J, Anderson C W, Appella E, Xu Y. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiscella M, Ullrich S J, Zambrano N, Shields M T, Lin D, Lees-Miller S P, Anderson C W, Mercer W E, Appella E. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- 31.Mayr G A, Reed M, Wang P, Wang Y, Schweds J F, Tegtmeyer P. Cancer Res. 1995;55:2410–2417. [PubMed] [Google Scholar]
- 32.Unger T, Sionov R V, Moallem E, Yee C L, Howley P M, Oren M, Haupt Y. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 33.Ashcroft M, Kubbutat M H, Vousden K H. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blattner C, Tobiasch E, Litfen M, Rahmsdorf H J, Herrlich P. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 35.Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Proc Natl Acad Sci USA. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Appella E, Anderson C W. Pathol Biol. 2000;48:227–245. [PubMed] [Google Scholar]
- 37.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 38.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]