Abstract
The essential oils of peppermint and spearmint are distinguished by the position of oxygenation on the constituent p-menthane monoterpenes. Peppermint produces monoterpenes bearing an oxygen at C3, whereas spearmint produces monoterpenes bearing an oxygen at C6. Branching of the monoterpene biosynthetic pathways in these species is determined by two distinct cytochrome P450s that catalyze the regiospecific hydroxylation of (−)-4S-limonene at C3 or C6 exclusively. cDNAs encoding the limonene-3-hydroxylase from peppermint and the limonene-6-hydroxylase from spearmint have been isolated, shown to be 70% identical at the amino acid level, and functionally expressed. A combination of domain swapping and reciprocal site-directed mutagenesis between these two enzymes demonstrated that the exchange of a single residue (F363I) in the spearmint limonene-6-hydroxylase led to complete conversion to the regiospecificity and catalytic efficiency of the peppermint limonene-3-hydroxylase.
Keywords: cytochrome P450, monoterpene, trans-carveol, trans-isopiperitenol, mint, Mentha
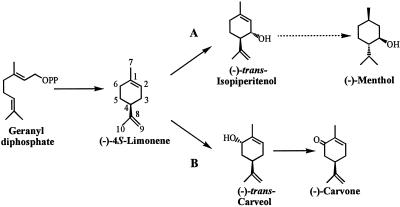
Spearmint (Mentha spicata) and peppermint (M. x piperita) are closely related mint species used for the commercial production of essential oils, the distinguishing characteristics of which are due to the position of oxygenation of the constituent p-menthane monoterpenes. Peppermint produces almost exclusively monoterpenes bearing an oxygen function at C3 [such as (−)-menthol responsible for the cooling sensation of peppermint], whereas spearmint produces almost exclusively monoterpenes bearing an oxygen function at C6 [such as (−)-carvone responsible for the typical spearmint note] (1) (Fig. 1). The regiospecificity of oxygenation is established very early in the monoterpene biosynthetic pathway in which the common olefinic precursor (−)-4S-limonene is hydroxylated exclusively at C3 to yield (−)-trans-isopiperitenol, which is subsequently transformed to the complex mixture of peppermint oil components (2). In spearmint, limonene is hydroxylated exclusively at C6 to yield (−)-trans-carveol and, following oxidation, (−)-carvone (2). These regiospecific hydroxylations are mediated by two distinct cytochrome P450 oxygenases that have been isolated from the epidermal oil glands of the respective mints and characterized (3).
Figure 1.
Pathways for monoterpene biosynthesis in peppermint (A) and spearmint (B). The dotted arrow indicates five enzymatic steps. OPP, diphosphate moiety.
Recently, a reverse genetic approach allowed isolation of a cDNA encoding the spearmint limonene-6-hydroxylase (designated SM12, assigned as CYP71D18) and two closely related cDNAs encoding the peppermint limonene-3-hydroxylase (PM2/CYP71D15 and PM17/CYP71D13) from the corresponding, highly enriched mint oil gland cDNA libraries, and all three clones were functionally expressed using the baculovirus–Spodoptera system (4). The two limonene-3-hydroxylases are 93% identical to each other in deduced amino acid sequence and exhibit 70% identity with the limonene-6-hydroxylase, and all three were assigned to the CYP71D subfamily of cytochrome P450 genes (5). Subsequent heterologous expression in Escherichia coli and Saccharomyces cerevisiae (6) has provided high yields of the functional enzymes to allow more detailed characterization of the mechanism and stereochemistry of the hydroxylation reactions.
Although many recombinant forms of eukaryotic P450s are now available, until recently this class of membrane proteins has proved to be recalcitrant to crystallization for x-ray structural determination. Much of what is known about cytochrome P450 structure–function relationships is therefore based on the soluble prokaryotic forms of the cytochrome for which x-ray crystal structures are available (7–11). Most approaches with eukaryotic P450 cytochromes have used chemical probes and mutagenesis studies that rest upon modeling from the soluble, bacterial forms (12–24). Although the sequence identity between any two P450s of defined three-dimensional structure reaches only 20%, the overall topology of the proteins is similar (25), and it now appears (26) that the structural fold is also similar for eukaryotic forms. The most dramatic variations between P450 primary structures are found in the regions responsible for substrate access and binding (27), a feature that often limits informative comparative study of selectivity and specificity with different, often phylogenetically distant, P450s. The close relationship between the (−)-limonene C3- and C6-hydroxylases offers a significant advantage in understanding the structural determinants of the regio and stereochemistry of oxygen insertion in P450 catalysis. In this paper, we describe the application of a systematic molecular strategy that has allowed the identification of a single amino acid residue responsible for the difference in the regiospecificity of these two enzymes.
Materials and Methods
Substrates, Reagents, and General Methods.
The sources of substrate and authentic standards, the optimized GC-based assay (with GC-MS verification), and protocols for quantification of cytochrome P450 by reduced CO-difference spectra, for determination of binding constants by ligand-induced difference spectra, and for SDS/PAGE and protein staining have been described (3, 4, 6). Restriction enzymes were from New England BioLabs, Pfu DNA polymerase was from Stratagene, and T4 DNA ligase was from GIBCO/BRL. The SM12 form of the spearmint limonene-6-hydroxylase was used and was compared with the PM2 isoform of the limonene-3-hydroxylase (4) as the latter consistently provided higher expression yields than did the PM17 isoform (6). All of the limonene hydroxylases were modified by substitution of the N terminus with the bovine 17α-hydroxylase sequence to optimize expression (6) in E. coli JM109 cells (Stratagene) using the pCWori(+) plasmid (28). The protocols for bacterial growth, induction, membrane isolation, and reconstitution of hydroxylase activity with purified, recombinant NADPH:cytochrome P450 reductase from spearmint have been reported in detail (6). For the determination of kinetic constants, the linear assays were standardized to the same level of membrane protein; the mean of triplicate measurements (±SE) are reported.
Construction of Hybrid cDNAs by Restriction Fragment Exchange.
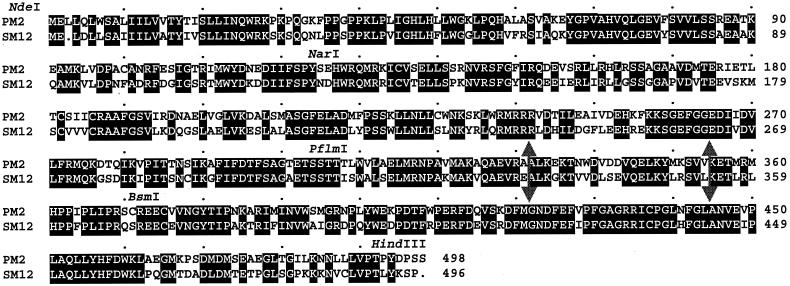
To target relevant domains, an initial set of chimeric cDNAs was constructed using the NdeI and the HindIII restriction sites introduced at the 5′- and the 3′-ends, respectively, of both hydroxylase cDNAs in combination with three naturally occurring, unique restriction sites located at the same position in the reading frame of both PM2 and SM12 sequences: NarI, located at codon 127 of SM12 and 128 of PM2; PflmI, located at codon 308 of SM12 and 309 of PM2; and BsmI, located at codon 375 of SM12 and 376 of PM2 (the numbering of the corresponding codons is slightly different in the two because of the presence of a gap in the membrane anchor region of SM12 when aligning both sequences) (Fig. 2). Because the pCWori(+) plasmid contains one PflmI and one BsmI restriction site, the hybrid cDNAs were constructed in pBluescript SK+ (pBSSK+) to simplify the restriction enzyme digestion before subcloning into the expression vector. The exchange of the four DNA fragments from both parent plasmids gave a combination of 14 hybrid cDNAs (Table 1). The ligations at the junctions were confirmed by redigestion, and the constructs were verified by restriction mapping before transfer, as NdeI–HindIII fragments, into the pCWori(+) expression plasmid.
Figure 2.
Alignment of deduced amino acid sequences of peppermint limonene-3-hydroxylase (PM2) and spearmint limonene-6-hydroxylase (SM12). Identical residues are indicated by boxing and reverse lettering. Positions of relevant restriction sites are indicated. The large double arrows delimit the three subdomains exchanged within the PflmI–BsmI domain.
Table 1.
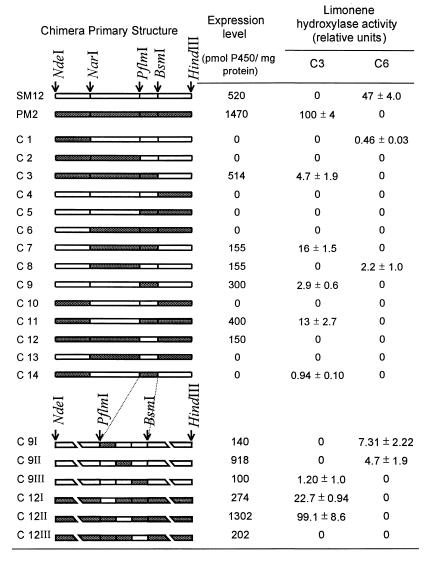
Schematic representation of the hydroxylase chimeras and their expression levels and activities
Chimeric cDNAs are represented by combination of white and gray boxes representing the wild-type SM12 (white) and PM2 (gray) coding sequences. The positions of the restriction sites are indicated by arrows. For the chimeras constructed by PCR, breaks have been introduced in the presentation to expand the region of interest. Expression levels were determined from the CO-difference spectra, and activity levels are relative to wild-type PM2 activity.
Construction of Hybrid cDNAs by PCR.
The exchange of smaller domains in the region between the PflmI and the BsmI restriction sites (Fig. 2) was performed by PCR using the overlap extension strategy (29). Three domains were exchanged: the first, containing the region from the PflmI site to codon 330 (E) for SM12 and to codon 331 (A) for PM2; the second, the region from codon 331 (A) to codon 353 (L) for SM12 and from codon 332 (A) to codon 354 (V) for PM2; and the third, the region from codon 354 (K) for SM12 and from codon 355 (K) for PM2 to the BsmI site (Fig. 2). All hybrid PCR products were purified by gel electrophoresis on 1% agarose, digested with NdeI and HindIII, and cloned directly into the pCWori(+) expression plasmid. The inserts were entirely sequenced to confirm that no unwanted alterations had occurred in the coding regions.
Site-Directed Mutagenesis.
Point mutations were introduced in the hydroxylase cDNAs by two PCR steps using one forward and one reverse mutagenic primer for each site-directed mutant. The modified cDNAs were then subcloned as NdeI–HindIII fragments directly into the pCWori(+) expression plasmid. Coding regions were fully sequenced to verify that no undesired mutations had been introduced by PCR.
Sequence Alignment and Modeling.
SM12 and PM2 protein sequences were aligned using GAP from the Wisconsin GCG Package (30) with standard defaults. For modeling, SM12 and PM2 protein sequences were aligned with sequences of P450cam (7), P450BM3 (8), and P450eryF (10) of defined crystal structure using GAP; subsequent corrections were made manually on the basis of the alignment described by Hasemann and coworkers (25). Models of PM2 and SM12 were then generated by automated molecular modeling (modeller 3 software) (31) using, as template, the three crystal structures of the soluble bacterial enzymes indicated above.
Results
Exchange of Large Restriction Fragments.
A domain-swapping strategy was first used in an attempt to localize regions of the proteins responsible for the control of regiospecificity in the hydroxylation of (−)-limonene. The PM2 and SM12 cDNAs contain three naturally occurring, unique restriction sites located in the same reading frame position (Fig. 2). Therefore, a first set of chimeric cDNAs was constructed using these three common sites to permit the exchange of the four derived DNA fragments. This approach yielded a combination of 14 different hybrid cDNAs (Table 1), all of which were cloned into pCWori(+) and transformed into E. coli JM109 cells for expression. Following induction and isolation of the membrane fractions, the content of functional cytochrome P450 was determined by CO-difference spectrometry to evaluate the presence of an appropriate heme environment, and each preparation was analyzed by SDS/PAGE followed by silver staining of the total membrane proteins to monitor expression of the corresponding 52-kDa recombinant protein (Table 1). Cytochrome P450 expression levels were generally lower for the chimeras than for the parental wild types, and in many cases the chimeric constructs did not afford a detectable difference spectrum. The results of SDS/PAGE were consistent with the spectrophotometric results, in that constructs that did not yield a functional cytochrome by CO-difference spectra also did not yield a detectable expressed protein at 52 kDa (data not shown). The apparent overall low expression of these chimeric cytochrome P450 constructs in E. coli may reflect perturbation in the folding and/or membrane integration of the corresponding proteins, leading to rapid degradation in the host; this also suggests that the two parent cytochromes differ in folding and stabilization processes. Thus, for example, chimeras 3 and 7 were reasonably well expressed, but the reciprocal constructs, chimeras 4 and 10, were not (Table 1).
Membrane protein preparations derived from each construct, as well as the parental P450s, were reconstituted using recombinant spearmint NADPH:cytochrome P450 reductase and tested for (−)-limonene hydroxylation activity using a sensitive GC-based assay (6). Seven of these preparations did not exhibit hydroxylase activity, but hydroxylase activity was readily detected for the remaining half, although the specific activity was low (ranging from 1 to 16% of that of the peppermint C3-hydroxylase) (Table 1). Except for chimera 12, one or the other regiospecific limonene hydroxylase activities was observed for all chimeras that yielded a reduced CO-difference spectrum. Additionally, hydroxylase activity was detected for chimeras 1 and 14, which afforded no CO-difference spectrum or visible recombinant protein, attesting to the sensitivity of the enzymatic assay. This initial effort, involving the exchange of four large domains, provided seven catalytically active limonene hydroxylases (chimeras 1, 3, 7, 8, 9, 11, and 14) from which a clear relationship between regiospecificity and primary structure could be established.
At the limit of sensitivity of the enzymatic assay, no mixtures of C3- and C6-hydroxylation products were observed, no products exhibiting hydroxylation at any other position of (−)-limonene were detected, and no alterations in the stereochemistry of hydroxylation were noted. Thus, all of the functional chimeric P450s conserve both regiospecificity and stereospecificity in demonstrating either C3-trans- or C6-trans-hydroxylation. Of the seven functional chimeras, five (chimeras 3, 7, 9, 11, and 14) exhibited C3-hydroxylation (yielding trans-isopiperitenol as the only product). The only domain originating from the peppermint (−)-limonene-3-hydroxylase, and common to these five chimeras, is the PflmI–BsmI domain. Notably, chimera 9 contains only this exchanged domain in a scaffold otherwise derived entirely from the spearmint (−)-limonene-6-hydroxylase (Table 1). Thus, the regiospecificity for (−)-limonene-3-hydroxylation is related to the peppermint cytochrome P450 PflmI–BsmI domain, the presence of which is necessary and sufficient to impart regiospecific (and stereospecific) C3-hydroxylase activity.
Only two chimeras (chimeras 1 and 8) yielded functional limonene-6-hydroxylase activity, and both of these contained the equivalent PflmI–BsmI domain from the spearmint C6-hydroxylase, suggesting that control of C6-regiospecificity is also related to this region. However, the fact that fewer chimeras were able to hydroxylate (−)-limonene at C6 than at C3, and that chimera 12 (the reciprocal of chimera 9) gave no activity (despite observable difference spectrum and visible recombinant protein) suggested that the presence of the spearmint PflmI–BsmI region is necessary but not sufficient in itself to direct C6-hydroxylation, or alternatively, that perturbations induced by exchange of large domains prevent folding to catalytically competent forms.
Exchange of Subdomains in the PflmI–BsmI Region.
The PflmI–BsmI region codes for 67 amino acids with 20 differences between the two parent enzymes (Fig. 2). This region was therefore divided into three subdomains with the expectation that exchange of these smaller segments would perturb protein folding to a lesser extent and allow more precise localization of the segment controlling regiospecificity of the two enzymes. A PCR strategy was used to construct six new chimeric cDNAs by exchanging three small subdomains, each coding for 21 to 23 residues and spanning the PflmI–BsmI region. As before, these new constructs were expressed in E. coli, the membrane fractions were isolated, and the cytochromes were characterized. The expression of this set of chimeric proteins was higher than for the previous set in which the modifications were more substantial (Table 1), and all yielded definitive CO-difference spectra with some approaching the specific content of the parental wild types. All of the new chimeras, except chimera 12III, exhibited (−)-limonene hydroxylase activity with specific activities ranging from 1 to 99% of the limonene-3-hydroxylase activity (Table 1). As before, the new chimeras were completely specific for C3-trans- or C6-trans-hydroxylation, and no alteration in stereochemistry or mixtures of products was observed.
For chimeras 9I and 9II, the introduction of the first two subdomains from the peppermint enzyme into the spearmint enzyme did not change the C6-regiospecificity of the parent hydroxylase (as did the introduction of the PflmI–BsmI domain for the original chimera 9). Thus, the first two subdomains do not contain residues responsible for the previously observed conversion in regiospecificity. For chimera 9III, however, the introduction of the third peppermint subdomain clearly altered regiochemistry in changing the spearmint limonene-6-hydroxylase to a limonene-3-hydroxylase.
The first two constructs derived from chimera 12 afforded relatively high specific activity for C3-hydroxylation like the parent peppermint enzyme. Thus, the first two subdomains are not responsible for the lack of activity of the original chimera 12, and these results confirm those obtained with chimeras 9I and 9II (i.e., that the first two subdomains of the PflmI–BsmI fragment do not influence regiospecificity). In the case of chimera 12III, the transfer of the third subdomain from SM12 into PM2 gave an inactive protein. This absence of activity was not due to misfolding, as a functional protein was expressed that gave a clear CO-difference spectrum. Thus, the reciprocal exchange of the third small subdomain led to complete conversion of C6 to C3 regioselectivity in the case of chimera 9III and to complete loss of activity in the case of chimera 12III with the C3 hydroxylase scaffold. Therefore, it can be concluded that the third subdomain of the PflmI–BsmI region of these enzymes is involved in substrate binding and likely contains residue(s) that influence the regiospecificity of limonene hydroxylation.
Reciprocal Site-Directed Mutagenesis.
There are only five amino acid differences in the third subdomain of the PflmI–BsmI region (Fig. 2). To define which of these are involved in substrate-binding interactions, the reciprocal exchanges were made in each parent cytochrome, and the five single mutant proteins for SM12 and for PM2 were expressed and evaluated (Table 2). All of the mutants were efficiently expressed in E. coli, and, for most, the specific limonene hydroxylase activity was in the range of the wild-type enzymes. As observed previously for all of the other hybrid enzymes, the nine catalytically active P450s were totally regioselective for either C3 or C6 hydroxylation; only PM2 mutant I364F expressed no detectable limonene hydroxylase activity.
Table 2.
Point mutants of the hydroxylases and their expression levels and activities
| Expression level, pmol P450/mg protein | Limonene hydroxylase activity, relative units
|
||
|---|---|---|---|
| C3 | C6 | ||
| SM12 wild type | 520 | 0 | 52.0 ± 5.4 |
| PM2 wild type | 1470 | 100 | 0 |
| SM12 L357M | 328 | 0 | 31.0 ± 0.4 |
| SM12 L359M | 613 | 0 | 42.3 ± 5.5 |
| SM12 F363I | 460 | 100 ± 15 | 0 |
| SM12 Q369S | 270 | 0 | 49.5 ± 1.6 |
| SM12 S370C | 614 | 0 | 56 ± 12 |
| PM2 M358L | 1090 | 100 ± 18 | 0 |
| PM2 M360L | 1300 | 7.5 ± 7.2 | 0 |
| PM2 I364F | 375 | 0 | 0 |
| PM2 S370Q | 884 | 98.2 ± 2.5 | 0 |
| PM2 C371S | 410 | 86.9 ± 24.0 | 0 |
Expression levels were determined from CO-difference spectra, and activity levels are relative to wild-type PM2 activity.
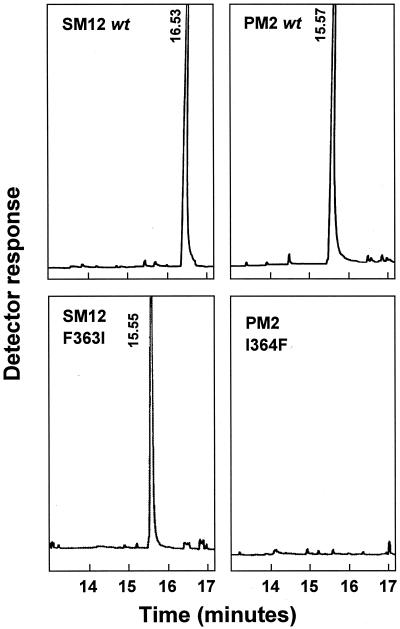
For the peppermint hydroxylase mutants and the spearmint hydroxylase mutants, swapping of the first two residues and the last two residues of the target segment did not influence the hydroxylation regiospecificity of the parental enzyme. By marked contrast, swapping the central residue in SM12 (the F363I mutation) led to complete conversion of the regioselectivity of this spearmint C6-hydroxylase to that of a peppermint-type limonene-3-hydroxylase (Fig. 3). Additionally, the SM12 F363I mutant protein displayed a catalytic rate 2-fold higher than the wild-type SM12 enzyme, and that reached the catalytic rate of the wild-type PM2 enzyme (Table 2). However, the reciprocal mutant in PM2 (i.e., I364F) was, at the limit of detection, inactive for (−)-limonene hydroxylation (Fig. 3). Although the expression level of the PM2 I364F mutant was four times lower than the PM2 wild type, a typical CO-difference spectrum was nevertheless obtained, indicating an appropriate heme environment.
Figure 3.
GC profiles of the hydroxylated products of (−)-limonene generated by wild-type (wt) cytochrome P450 SM12 and PM2, and mutants SM12 F363I and PM2 I364F, when reconstituted with purified spearmint NADPH:cytochrome P450 reductase. The products identified are (−)-trans-carveol (16.53 min) and (−)-trans-isopiperitenol (15.56 ± 0.01 min).
A likely explanation for the lack of activity of PM2 I364F is that the mutation adversely influences substrate binding. The binding behavior of SM12, PM2, SM12 F363I, and PM2 I364F was therefore examined by evaluating the spectral shift accompanying the change in low to high spin state induced by substrate displacement of water as the sixth ligand to the heme iron (32). The spectral shift was easily detected as a type I spectrum (maximum ∼390 nm and minimum ∼420 nm) (33), from which the plot of absorbance difference vs. substrate concentration allowed determination of the apparent dissociation constant (Ks,app). For the wild-type enzymes, the Ks,app values were 3.8 ± 0.9 μM and 0.66 ± 0.07 μM for SM12 and PM2, respectively. For the SM12 F363I mutant, Ks,app was 2.6 ± 0.3 μM, and so slightly lower than for the corresponding wild type. Limonene-induced difference spectra were also obtained for the PM2 I364F mutant, but substantially higher substrate concentrations were required, leading to an estimated Ks,app value of 94 ± 44 μM (the error values reflect limitations in obtaining precise concentrations for a substrate that is both volatile and of quite limited solubility in water). Thus, in the PM2 I364F mutant, the binding affinity for (−)-limonene is adversely affected, and the binding orientation of this substrate is also likely so compromised as to prevent catalysis.
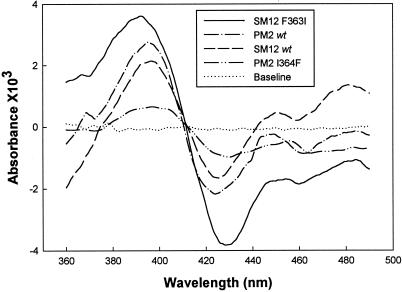
The amplitude of the type I spectrum induced by a saturating concentration of (−)-limonene (ΔAmax) is dependent on the spin ratio of the P450 in the absence of substrate and the ability of the substrate to efficiently displace the water molecule(s) from the active site pocket. The ΔAmax values were quite different for the two wild-type enzymes and their respective point mutants (Fig. 4). By normalizing to the amount of P450 (ΔAmax/mmol P450), the highest amplitude was obtained with the SM12 F363I mutant (78A/mmol), a value twice that of the SM12 parent (38 A/mmol) that indicates improved access to the active site by the isoleucine for phenylalanine substitution. In the case of PM2 and the I364F mutant, the normalized values were lower than for the SM12 scaffold (suggestive of more limited active site access) but not significantly different from each other (17 and 19 A/mmol, respectively), indicating little direct influence by the mutation.
Figure 4.
(−)-Limonene-induced binding spectra obtained with membranous preparations of SM12, PM2, SM12 F363I, and PM2 I364F. Spectra were recorded at 0.2 mg of protein/ml (corresponding to 92, 294, 104, and 75 pmol of P450 for SM12 F363I, PM2, SM12, and PM2 I364F, respectively) with 100 μM (−)-limonene (for PM2 I364F, the limonene concentration was 200 μM).
Discussion
The reciprocal exchange of amino acids has been used to identify residues involved in substrate binding of related P450s (14–17) that can display broad and overlapping specificities. There are several examples in which minor alteration in the size of an amino acid chain influences P450 catalysis (17–22), but multiple mutations are often required to substantially modify activity (14, 15, 17, 23). In the present case, reciprocal exchange between two related (70% sequence identity) regiospecific P450 limonene hydroxylases from spearmint and peppermint allowed identification of a single residue that is seemingly critical for the control of substrate-binding orientation. Thus, the F363I mutation in the C6-hydroxylase led to complete conversion to a C3-hydroxylase with excellent catalytic efficiency. The reciprocal I364F modification in the C3-hydroxylase led to inactive enzyme despite the fact that the protein was properly folded as evidenced by the CO-binding spectrum. Because both wild-type P450s, and all active chimeras and mutants tested, seem to provide only one or the other specificities (C3-trans or C6-trans), the active sites must be sufficiently constrained to permit only one or the other substrate orientations with respect to the heme iron. The catalytic failure of the I364F modification could reflect steric obstruction to productive binding or simply access, for which the significant increase in Ks,app for this mutant is suggestive.
The purified SM12 and PM2 enzymes are always found in mixed spin state and cannot be saturated by (−)-limonene (data not shown), indicating limited solvent and substrate access to the active site. The increased affinity for limonene and the increased amplitude of the binding spectrum for the regiochemically altered SM12 F363I mutant indicate improved access compared with the wild type, an observation consistent with the size reduction of the side chain and concomitant increase of the internal active site volume. The amplitude of the binding spectrum for the PM2 wild type is lower by a factor of two compared with SM12, suggesting that the active site of this wild-type C3-hydroxylase is substantially more restricted. The introduction of increased steric bulk in the PM2 I364F mutant seems to sufficiently perturb the already limited access to prevent catalysis, thereby obscuring any alteration in substrate-binding orientation that might otherwise be reflected in modified reaction stereochemistry.
Comparison of crystal structures of soluble, prokaryotic cytochrome P450s has shown similar secondary structural elements and overall folding patterns despite the low primary sequence similarities (25, 26, 34), and sequence comparison with several families of eukaryotic P450s (25, 26) and the structure of the first crystallized microsomal P450 (35) suggest a common structural fold for all eukaryotic and prokaryotic forms. There are many well-conserved structural elements related to oxygen binding and activation that are common to all P450s (25, 26, 36); however, the regions of the protein involved in substrate-binding interactions are structurally quite variable in reflecting the great diversity in substrate and product specificity of the P450 superfamily. Alignment studies by Gotoh (27) have defined six putative substrate recognition sites (SRS) that occur in common regions of primary sequence for prokaryotic and eukaryotic P450s (25). Site-directed mutagenesis studies with CYP2 family members and other mammalian P450 families (12–19, 26) have largely supported this concept by localizing, in the predicted SRS, the contact residues that direct substrate specificity and product outcome. Sequence alignments in the present instance reveal that the residues Phe-363 and Ile-364 of SM12 and PM12, respectively, reside in SRS-5 described by Gotoh (27).
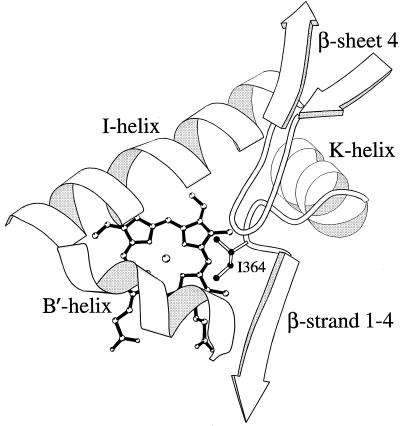
SRS-5 in structurally defined cytochrome P450s is localized in a region extending from the end of the K helix to the beginning of β-strand 1–4 and delimits one side of the substrate pocket (27). Directed mutations in this region have been reported to influence substrate selectivity and product specificity for several P450s (14, 19–22). To understand the potential role of Phe-363 and Ile-364 in the active sites of the spearmint and peppermint limonene hydroxylases, models of the two proteins were constructed based upon the scaffolds of the crystalline prokaryotic P450s. These models are consistent with the experimental data in indicating that the Phe-363 and Ile-364 residues protrude into the active site on one side of the pocket (Fig. 5) and therefore can interact directly with the substrate to influence binding orientation. Substrate docking in the model will be required to further define the means by which altered substrate binding promotes C3-hydroxylation by the SM12 F363I mutant. Inspection of the models also indicates structural differences between the two active sites that may underlie the catalytic failure of the PM2 I364F mutant and suggests other regions of the active site that may require modification to both restore activity and alter regiospecificity of the resulting protein. These targeted regions include the center of the I-helix (SRS-4) and the B′-helix region (SRS-1), which delimit the sides of the pocket opposite to SRS-5, and the β-turn at the end of β-sheet 4 (SRS-6), which extends the active site on the same side and over SRS-5 (Fig. 5).
Figure 5.
Active-site model of PM2 illustrating the side chain of Ile-364 and the heme in ball and stick form and the denoted protein domains surrounding the active site as backbones. The image was created with the molscript program (37).
Acknowledgments
We thank C. Sanchez, G. Munske, and D. Pouchnik for nucleotide sequencing, Dr. David Hyatt for assistance with the graphics, and Professor Jeffrey Jones for assistance with the molecular modeling. This investigation was supported by a grant from the National Science Foundation.
Abbreviation
- SRS
substrate recognition site(s)
References
- 1.Lawrence B M. In: Essential Oils. Mookherjee B D, Mussinan C J, editors. Wheaton, IL: Allured Press; 1981. pp. 1–81. [Google Scholar]
- 2.Croteau R, Gershenzon J. Recent Adv Phytochem. 1994;28:193–229. [Google Scholar]
- 3.Karp F, Mihaliak C A, Harris J L, Croteau R. Arch Biochem Biophys. 1990;276:219–226. doi: 10.1016/0003-9861(90)90029-x. [DOI] [PubMed] [Google Scholar]
- 4.Lupien S, Karp F, Wildung M, Croteau R. Arch Biochem Biophys. 1999;368:181–192. doi: 10.1006/abbi.1999.1298. [DOI] [PubMed] [Google Scholar]
- 5.Nelson D R, Koymans L, Kamataki T, Stegeman J J, Feyereisen R, Waxman D J, Waterman M R, Gotoh O, Coon M J, Estabrook R J, et al. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Haudenschild C, Schalk M, Karp F, Croteau R. Arch Biochem Biophys. 2000;379:127–136. doi: 10.1006/abbi.2000.1864. [DOI] [PubMed] [Google Scholar]
- 7.Poulos T L, Finzel B C, Howard A J. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 8.Ravichandran K G, Boddupalli S S, Hasemann C A, Peterson J A, Deisenhofer J. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 9.Hasemann C A, Ravichandran K G, Peterson J A, Deisenhofer J. J Mol Biol. 1994;236:1169–1185. doi: 10.1016/0022-2836(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 10.Cupp-Vickery J R, Poulos T L. Nat Struct Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 11.Park S Y, Shimizu H, Adachi S, Nakagawa A, Tanaka I, Nakahara K, Shoun H, Obayashi E, Nakamura H, Iizura T, et al. Nat Struct Biol. 1997;4:827–832. doi: 10.1038/nsb1097-827. [DOI] [PubMed] [Google Scholar]
- 12.von Wachenfeldt C, Johnson E F. In: Cytochrome P450: Structure, Mechanism and Biochemistry. 2nd Ed. Ortiz de Montellano P, editor. New York: Plenum Press; 1995. pp. 183–223. [Google Scholar]
- 13.Negishi M, Uno T, Darden T A, Sueyoshi T, Pedersen L G. FASEB J. 1996;10:683–689. doi: 10.1096/fasebj.10.7.8635685. [DOI] [PubMed] [Google Scholar]
- 14.He Y Q, Szklarz G D, Halpert J R. Arch Biochem Biophys. 1996;335:152–160. doi: 10.1006/abbi.1996.0493. [DOI] [PubMed] [Google Scholar]
- 15.Böttner B, Denner K, Bernhardt R. Eur J Biochem. 1998;252:458–466. doi: 10.1046/j.1432-1327.1998.2520458.x. [DOI] [PubMed] [Google Scholar]
- 16.Zimmer T, Scheller U, Takagi M, Schunck W-H. Eur J Biochem. 1998;256:398–403. doi: 10.1046/j.1432-1327.1998.2560398.x. [DOI] [PubMed] [Google Scholar]
- 17.Lindberg R L P, Negishi M. Nature (London) 1989;339:632–634. doi: 10.1038/339632a0. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki M, Darden T A, Pedersen L G, Negishi M. Biochemistry. 1995;34:5056–5059. doi: 10.1021/bi00015a016. [DOI] [PubMed] [Google Scholar]
- 19.Ellis S W, Rowland K, Ackland M J, Rekka E, Simula A P, Lennard M S, Wolf C R, Tucker G T. Biochem J. 1996;316:647–654. doi: 10.1042/bj3160647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanna I H, Roberts E S, Hollenberg P F. Biochemistry. 1998;37:311–318. doi: 10.1021/bi971528s. [DOI] [PubMed] [Google Scholar]
- 21.He Y A, He Y Q, Szklarz G D, Halpert J R. Biochemistry. 1997;36:8831–8839. doi: 10.1021/bi970182i. [DOI] [PubMed] [Google Scholar]
- 22.Richardson T H, Johnson E F. J Biol Chem. 1994;269:23937–23943. [PubMed] [Google Scholar]
- 23.He Y Q, Harlow G R, Szklarz G D, Halpert J R. Arch Biochem Biophys. 1998;350:333–339. doi: 10.1006/abbi.1997.0516. [DOI] [PubMed] [Google Scholar]
- 24.Dierks E A, Davis S C, Ortiz de Montellano P R. Biochemistry. 1998;37:1839–1847. doi: 10.1021/bi972458s. [DOI] [PubMed] [Google Scholar]
- 25.Hasemann C A, Kurumbail R G, Boddupalli S S, Peterson J A, Deisenhofer J. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 26.Graham S E, Peterson J A. Arch Biochem Biophys. 1999;369:24–29. doi: 10.1006/abbi.1999.1350. [DOI] [PubMed] [Google Scholar]
- 27.Gotoh O. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 28.Barnes H J. Methods Enzymol. 1996;272B:3–14. doi: 10.1016/s0076-6879(96)72003-7. [DOI] [PubMed] [Google Scholar]
- 29.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 30.Devereux J, Haeberlin P, Smithies O. Nucleic Acids Res. 1994;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sali A, Blundell T L. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 32.Raag R, Poulos T L. Biochemistry. 1989;28:917–922. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- 33.Jefcoate C R. Methods Enzymol. 1978;52:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- 34.Peterson J A, Graham-Lorence S E. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 2nd Ed. Ortiz de Montellano P R, editor. New York: Plenum Press; 1995. pp. 151–180. [Google Scholar]
- 35.Williams P A, Cosme J, Sridhar V, Johnson E F, McRee D E. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 36.Graham-Lorence S E, Peterson J A. Methods Enzymol. 1996;272B:315–326. doi: 10.1016/s0076-6879(96)72037-2. [DOI] [PubMed] [Google Scholar]
- 37.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]