Abstract
CASE REPORT—A patient is described who developed hepatitis B virus (HBV) reinfection five months following liver transplantation. Failure of hepatitis B immunoglobulin prophylaxis was associated with the emergence of mutations. HBV gene sequencing identified nucleotide substitutions associated with amino acid changes, one within the major hydrophilic region (MHR) of the HBV surface antigen at amino acid position 144 and one outside the MHR. Because of the overlapping reading frames of surface and polymerase genes, the latter surface antigen change was associated with an amino acid change in the polymerase protein. The patient developed significant allograft hepatitis and was treated with lamivudine (3TC) 100 mg daily. Rapid decline of serum HBV DNA was observed with loss of HBV e antigen and HBV surface antigen from serum. There was normalisation of liver biochemistry, and liver immunohistochemistry showed a reduction in HBV core and disappearance of HBs antigen staining. CONCLUSION—Surface antigen encoding gene mutations associated with HBIg escape may be associated with alteration of the polymerase protein. The polymerase changes may affect sensitivity to antiviral treatment. Selection pressure on one HBV reading frame (for example, HBIg pressure on HBsAg, or nucleoside analogue pressure on polymerase protein) may alter the gene product of the overlapping frame. Such interactions are relevant to strategies employing passive immune prophylaxis and antiviral treatment. Keywords: liver transplantation; prophylaxis; escape mutants; lamivudine
Full Text
The Full Text of this article is available as a PDF (114.4 KB).
Figure 1 .
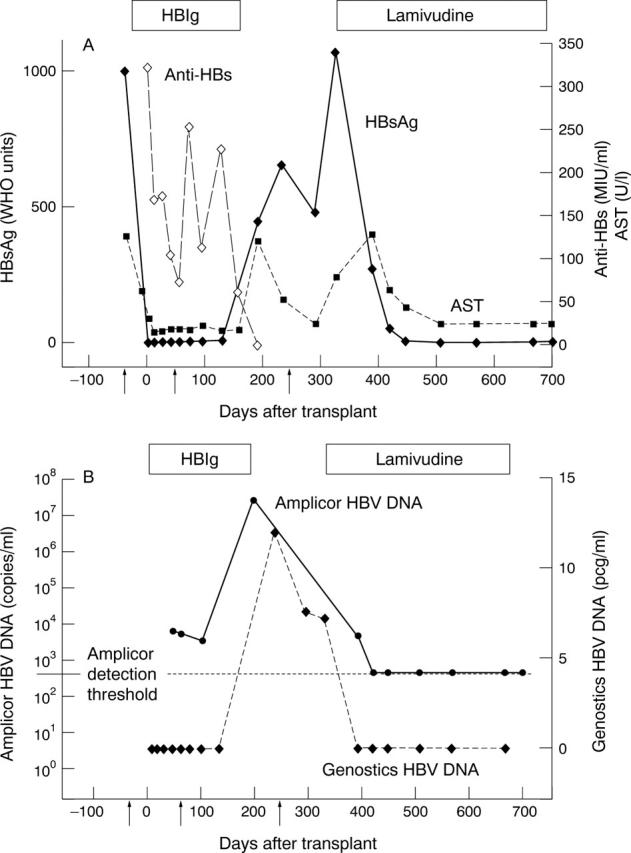
Serum levels of HBsAg, anti-HBs, and AST (A) and levels of HBV DNA (B) prior to liver transplantation, during HBIg prophylaxis, and following recurrence of infection. Arrows show time points of HBV surface gene sequence analysis.
Figure 2 .
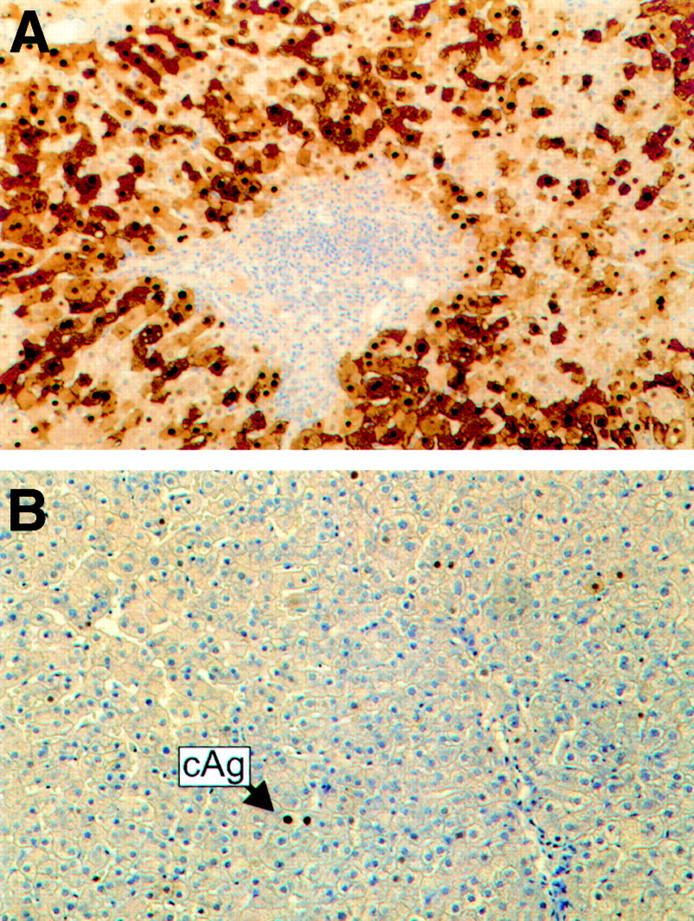
(A) Hepatitis B core (HBc) staining of liver biopsy taken 10 months post-liver transplantation, following the development of allograft reinfection. Portal tract contains a moderately dense infiltrate of lymphocytes. There is widespread nuclear and cytoplasmic immunostaining for HBcAg (immunoperoxidase stain). (B) Biopsy 16 months post-liver transplantation and after four months of lamivudine therapy (100 mg daily). Immunostaining for HBcAg is now restricted to the nuclei of occasional hepatocytes (cAg). Little inflammatory activity is present.
Figure 3 .
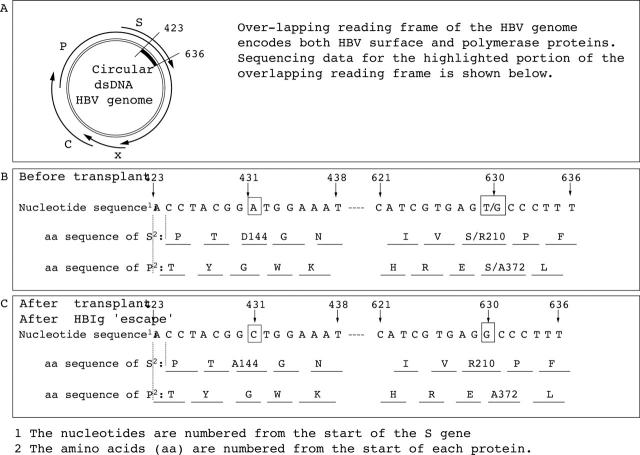
(A) The HBV genome encodes four proteins (core (C), surface (S), polymerase (P), and X proteins). The overlapping reading frame nature of the HBV genome allows a mutation within the genome encoding S and P proteins potentially to alter the amino acid sequence of the S and P proteins. Sequence data show nucleotide and amino acid sequences of the hepatitis B surface gene (nucleotides 423-438 and 621-636) and the corresponding sequence of HBV DNA polymerase: (B) before liver transplantation and (C) after "viral escape" from hepatitis B immunoglobulin prophylaxis.