Abstract
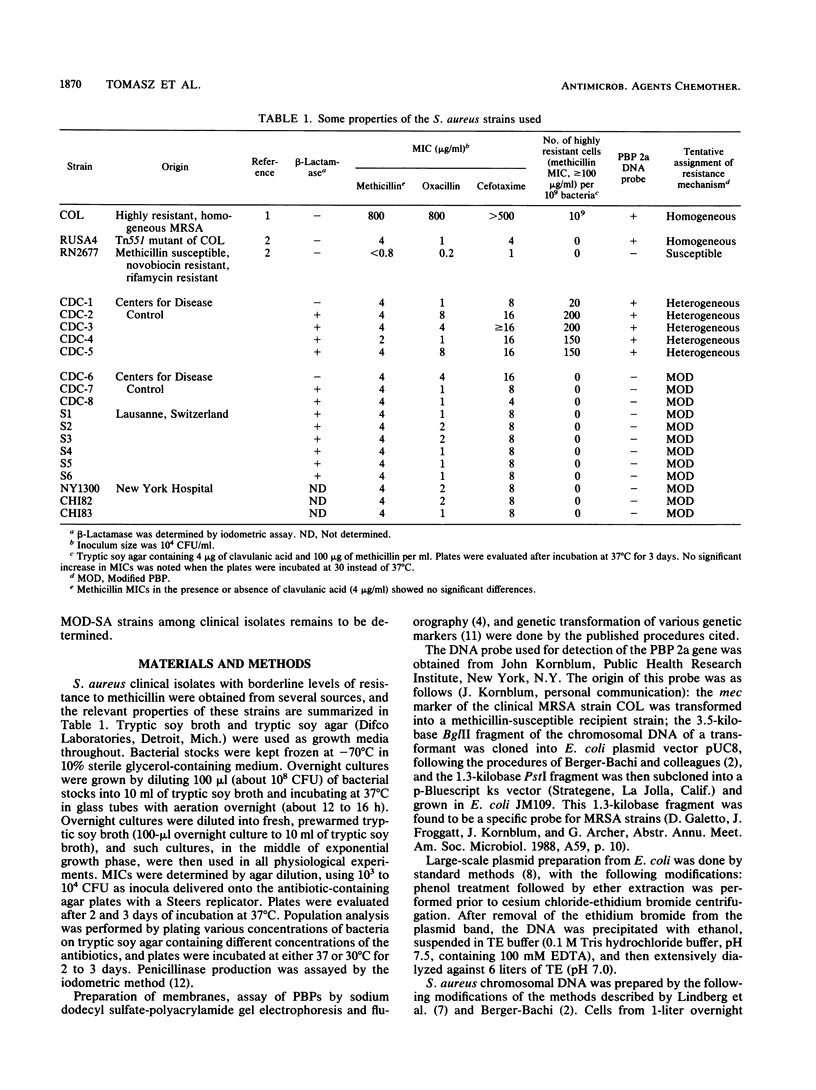
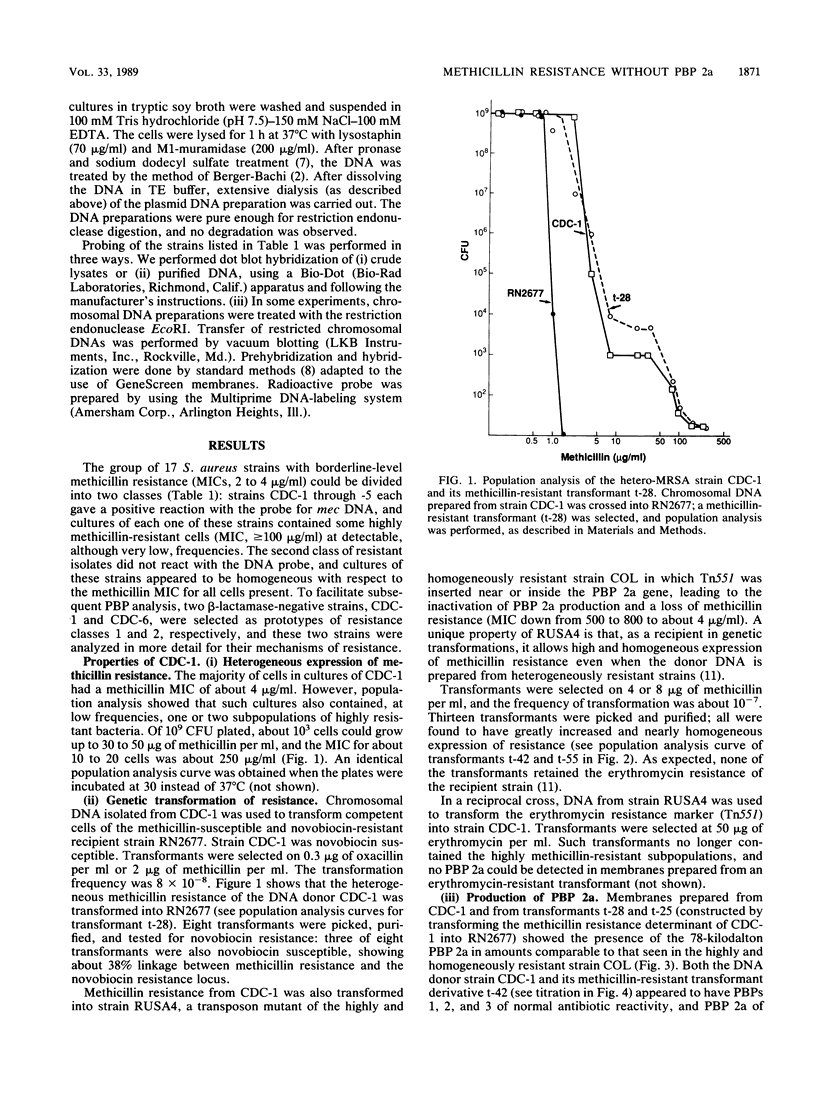
Seventeen clinical isolates of Staphylococcus aureus (from the United States and Europe) selected for low (borderline)-level methicillin resistance (MIC of methicillin, 2 to 4 micrograms/ml; MIC of oxacillin, 0.5 to 8 micrograms/ml) were examined for their mechanisms of resistance. Five strains were typical of heterogeneous S. aureus: they gave positive reactions with a DNA probe specific for mec and contained a small fraction (10(-6] of highly resistant cells (MIC, greater than 100 micrograms/ml). The rest of the 12 strains were homogeneous with respect to their methicillin resistance: the MIC of methicillin for all cells was 2 to 4 micrograms/ml, and no cells for which MICs were 50 micrograms/ml or higher were detectable (less than 10(-9]. None of these strains reacted with the mec-specific DNA probe. One representative strain of each group was characterized in more detail. Strain CDC-1, prototype of heterogeneous methicillin-resistant S. aureus, contained penicillin-binding protein (PBP) 2a; its DNA could transform a methicillin-susceptible and novobiocin-resistant recipient to methicillin resistance with ca. 35% linkage to Novr. Introduction of the "factor X" determinant (K. Murakami and A. Tomasz, J. Bacteriol. 171:874-879, 1989) converted strain CDC-1 to high, homogeneous resistance. Strain CDC-6, prototype of the second group of isolates, showed completely homogeneous MICs of methicillin, oxacillin, and cefotaxime. The strain contained modified "normal" PBPs: PBPs 1 and 2 showed low drug reactivity (and/or cellular amounts), and PBP 4 was present in elevated amounts. No PBP 2a could be detected. DNA isolated from strain CDC-6 could transform the methicillin-susceptible and novobiocin-resistant strain to methicillin resistance in a multistep fashion, but this resistance showed no genetic linkage to the Nov marker. We suggest that staphylococci with borderline resistance may contain at least three different classes of mechanism: heterogeneous, methicillin-resistant S. aureus, PBPs of modified drug reactivities, and the previously reported hyperproduction of beta-lactamase (L.K. McDougal and C. Thornsberry, J. Clin Microbiol. 23:832-839, 1986).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck W. D., Berger-Bächi B., Kayser F. H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986 Feb;165(2):373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J Bacteriol. 1983 Apr;154(1):479–487. doi: 10.1128/jb.154.1.479-487.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. F., Reynolds P. E. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 1980 Dec 29;122(2):275–278. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Jan;29(1):85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984 May;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis B., Matthews P. R., Stewart P. R. The expression in Staphylococcus aureus of cloned DNA encoding methicillin resistance. J Gen Microbiol. 1988 Jun;134(6):1465–1469. doi: 10.1099/00221287-134-6-1465. [DOI] [PubMed] [Google Scholar]
- Lindberg M., Sjöström J. E., Johansson T. Transformation of chromosomal and plasmid characters in Staphylococcus aureus. J Bacteriol. 1972 Feb;109(2):844–847. doi: 10.1128/jb.109.2.844-847.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M., Song M. D., Ishino F., Wachi M., Doi M., Inoue M., Ubukata K., Yamashita N., Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986 Sep;167(3):975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal L. K., Thornsberry C. The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J Clin Microbiol. 1986 May;23(5):832–839. doi: 10.1128/jcm.23.5.832-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOVICK R. P. Staphylococcal penicillinase and the new penicillins. Biochem J. 1962 May;83:229–235. doi: 10.1042/bj0830229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonin E., Tomasz A. Beta-lactam-specific resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Oct;30(4):577–583. doi: 10.1128/aac.30.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trees D. L., Iandolo J. J. Identification of a Staphylococcus aureus transposon (Tn4291) that carries the methicillin resistance gene(s). J Bacteriol. 1988 Jan;170(1):149–154. doi: 10.1128/jb.170.1.149-154.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Yamashita N., Konno M. Occurrence of a beta-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1985 May;27(5):851–857. doi: 10.1128/aac.27.5.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zighelboim S., Tomasz A. Penicillin-binding proteins of multiply antibiotic-resistant South African strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980 Mar;17(3):434–442. doi: 10.1128/aac.17.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]






