Abstract
BACKGROUND AND AIMS—Genetic predisposition for inflammatory bowel disease (IBD) has been demonstrated by epidemiological and genetic linkage studies. Genetic linkage of IBD to chromosome 3 has been observed previously. A high density analysis of chromosome 3p was performed to confirm prior linkages and elucidate potential genetic associations. METHODS—Forty three microsatellite markers on chromosome 3 were genotyped in 353 affected sibling pairs of North European Caucasian extraction (average marker density 2 cM in the linkage interval). Marker order was defined by genetic and radiation hybrid techniques. RESULTS—The maximum single point logarithm of odds (LOD) score was observed for Crohn's disease at D3S3591. Peak multipoint LOD scores of 1.65 and 1.40 for the IBD phenotype were observed near D3S1304 (distal 3p) and near D3S1283 in the linkage region previously reported. Crohn's disease contributed predominantly to the linkage. The transmission disequilibrium test showed significant evidence of association (p=0.009) between allele 4 of D3S1076 and the IBD phenotype (51 transmitted v 28 non-transmitted). Two known polymorphisms in the CCR2 and CCR5 genes were analysed, neither of which showed significant association with IBD. Additional haplotype associations were observed in the vicinity of D3S1076. CONCLUSIONS—This study provides confirmatory linkage evidence for an IBD susceptibility locus on chromosome 3p and suggests that CCR2 and CCR5 are unlikely to be major susceptibility loci for IBD. The association findings in this region warrant further investigation. Keywords: inflammatory bowel disease; fine mapping; chromosome 3
Full Text
The Full Text of this article is available as a PDF (155.5 KB).
Figure 1 .
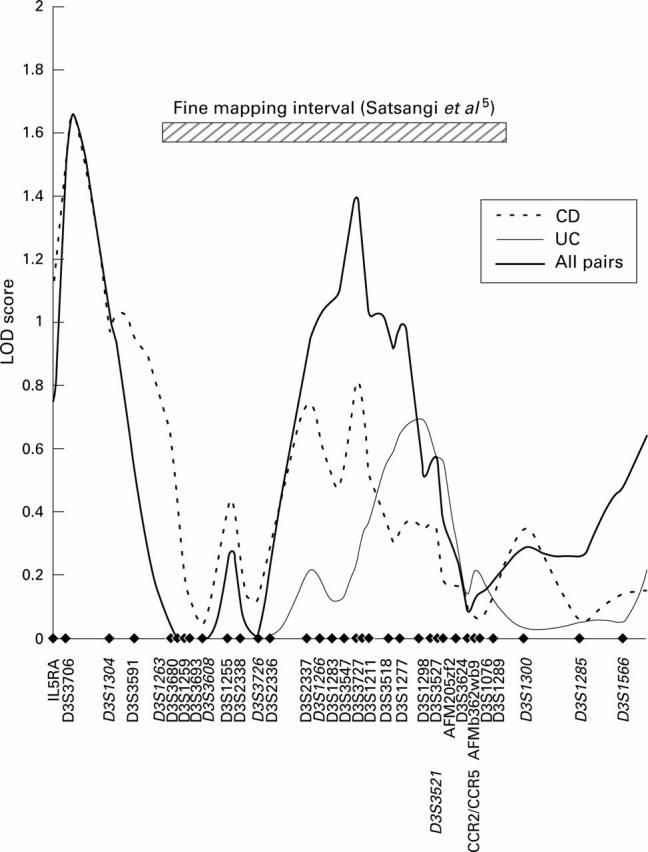
Multipoint MLS curves for the markers genotyped in the saturation region on chromosome 3p. Markers were genotyped in 353 affected sibling pairs and analysed for allele sharing using the "weighted pairs" option of Mapmaker/Sibs.31 Results for Crohn's disease (CD), ulcerative colitis (UC), and all pairs are shown. Genetic distances were estimated from the marker data using Multimap. The marker positions are indicated by filled diamonds. The markers also used in the genome wide analysis are indicated in italics.
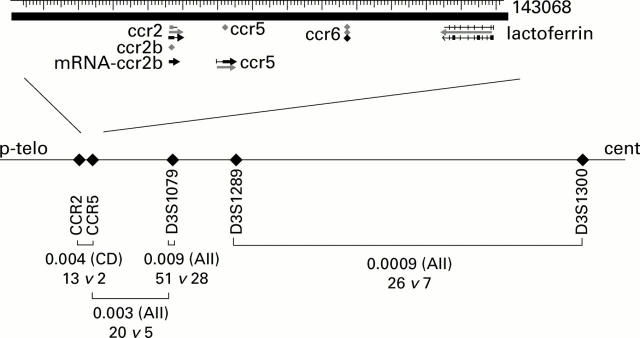
Figure 2 .
Detailed view of the putative association region around D3S1076. In the lower part of the figure, markers are arranged according to the genetic map. The brackets indicate single point and two point associations of markers. For each finding, the nominal p value with the phenotype category in which it was obtained and the number of transmitted versus non-transmitted alleles/haplotypes is indicated. The upper part of the figure offers a close up view of the CCR gene cluster and was derived from the NCBI website (www.ncbi.nlm.nih.gov, based on the Genbank record U95626).
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Annese V., Latiano A., Bovio P., Forabosco P., Piepoli A., Lombardi G., Andreoli A., Astegiano M., Gionchetti P., Riegler G. Genetic analysis in Italian families with inflammatory bowel disease supports linkage to the IBD1 locus--a GISC study. Eur J Hum Genet. 1999 Jul;7(5):567–573. doi: 10.1038/sj.ejhg.5200328. [DOI] [PubMed] [Google Scholar]
- Bals R., Weiner D. J., Moscioni A. D., Meegalla R. L., Wilson J. M. Augmentation of innate host defense by expression of a cathelicidin antimicrobial peptide. Infect Immun. 1999 Nov;67(11):6084–6089. doi: 10.1128/iai.67.11.6084-6089.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant S. R., Fu Y., Fields C. T., Baltazar R., Ravenhill G., Pickles M. R., Rohal P. M., Mann J., Kirschner B. S., Jabs E. W. American families with Crohn's disease have strong evidence for linkage to chromosome 16 but not chromosome 12. Gastroenterology. 1998 Nov;115(5):1056–1061. doi: 10.1016/s0016-5085(98)70073-3. [DOI] [PubMed] [Google Scholar]
- Cavanaugh J. A., Callen D. F., Wilson S. R., Stanford P. M., Sraml M. E., Gorska M., Crawford J., Whitmore S. A., Shlegel C., Foote S. Analysis of Australian Crohn's disease pedigrees refines the localization for susceptibility to inflammatory bowel disease on chromosome 16. Ann Hum Genet. 1998 Jul;62(Pt 4):291–298. doi: 10.1046/j.1469-1809.1998.6240291.x. [DOI] [PubMed] [Google Scholar]
- Cho J. H., Nicolae D. L., Gold L. H., Fields C. T., LaBuda M. C., Rohal P. M., Pickles M. R., Qin L., Fu Y., Mann J. S. Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q, and 4q: evidence for epistasis between 1p and IBD1. Proc Natl Acad Sci U S A. 1998 Jun 23;95(13):7502–7507. doi: 10.1073/pnas.95.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran M. E., Lau K. F., Hampe J., Schreiber S., Bridger S., Macpherson A. J., Cardon L. R., Sakul H., Harris T. J., Stokkers P. Genetic analysis of inflammatory bowel disease in a large European cohort supports linkage to chromosomes 12 and 16. Gastroenterology. 1998 Nov;115(5):1066–1071. doi: 10.1016/s0016-5085(98)70075-7. [DOI] [PubMed] [Google Scholar]
- Duerr R. H., Barmada M. M., Zhang L., Davis S., Preston R. A., Chensny L. J., Brown J. L., Ehrlich G. D., Weeks D. E., Aston C. E. Linkage and association between inflammatory bowel disease and a locus on chromosome 12. Am J Hum Genet. 1998 Jul;63(1):95–100. doi: 10.1086/301929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epplen C., Frank G., Gomolka M., Nagy M., Nürnberg P., Epplen J. T. Dinucleotide repeat polymorphism in the IL2 and IL5RA genes. Hum Mol Genet. 1994 Apr;3(4):679–679. doi: 10.1093/hmg/3.4.679-a. [DOI] [PubMed] [Google Scholar]
- Hampe J., Hermann B., Bridger S., MacPherson A. J., Mathew C. G., Schreiber S. The interferon-gamma gene as a positional and functional candidate gene for inflammatory bowel disease. Int J Colorectal Dis. 1998;13(5-6):260–263. doi: 10.1007/s003840050173. [DOI] [PubMed] [Google Scholar]
- Hampe J., Schreiber S., Shaw S. H., Lau K. F., Bridger S., Macpherson A. J., Cardon L. R., Sakul H., Harris T. J., Buckler A. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet. 1999 Mar;64(3):808–816. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J., Shaw S. H., Saiz R., Leysens N., Lantermann A., Mascheretti S., Lynch N. J., MacPherson A. J., Bridger S., van Deventer S. Linkage of inflammatory bowel disease to human chromosome 6p. Am J Hum Genet. 1999 Dec;65(6):1647–1655. doi: 10.1086/302677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J., Wienker T., Nürnberg P., Schreiber S. Mapping genes for polygenic disorders: considerations for study design in the complex trait of inflammatory bowel disease. Hum Hered. 2000 Mar-Apr;50(2):91–101. doi: 10.1159/000022896. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Holmans P. Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet. 1993 Feb;52(2):362–374. [PMC free article] [PubMed] [Google Scholar]
- Hugot J. P., Laurent-Puig P., Gower-Rousseau C., Olson J. M., Lee J. C., Beaugerie L., Naom I., Dupas J. L., Van Gossum A., Orholm M. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996 Feb 29;379(6568):821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- Idury R. M., Cardon L. R. A simple method for automated allele binning in microsatellite markers. Genome Res. 1997 Nov;7(11):1104–1109. doi: 10.1101/gr.7.11.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S. V., Izotova L. S., Mirochnitchenko O. V., Lee C., Pestka S. The intracellular domain of interferon-alpha receptor 2c (IFN-alphaR2c) chain is responsible for Stat activation. Proc Natl Acad Sci U S A. 1999 Apr 27;96(9):5007–5012. doi: 10.1073/pnas.96.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L., Daly M. J., Reeve-Daly M. P., Lander E. S. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996 Jun;58(6):1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L., Lander E. S. Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet. 1995 Aug;57(2):439–454. [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L., Lander E. S. Limits on fine mapping of complex traits. Am J Hum Genet. 1996 May;58(5):1092–1093. [PMC free article] [PubMed] [Google Scholar]
- Lander E., Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995 Nov;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lennard-Jones J. E. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2–19. doi: 10.3109/00365528909091339. [DOI] [PubMed] [Google Scholar]
- Lewis A. J., Manning A. M. New targets for anti-inflammatory drugs. Curr Opin Chem Biol. 1999 Aug;3(4):489–494. doi: 10.1016/S1367-5931(99)80071-4. [DOI] [PubMed] [Google Scholar]
- Liu R., Paxton W. A., Choe S., Ceradini D., Martin S. R., Horuk R., MacDonald M. E., Stuhlmann H., Koup R. A., Landau N. R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996 Aug 9;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Matise T. C., Perlin M., Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994 Apr;6(4):384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- Ohmen J. D., Yang H. Y., Yamamoto K. K., Zhao H. Y., Ma Y., Bentley L. G., Huang Z., Gerwehr S., Pressman S., McElree C. Susceptibility locus for inflammatory bowel disease on chromosome 16 has a role in Crohn's disease, but not in ulcerative colitis. Hum Mol Genet. 1996 Oct;5(10):1679–1683. doi: 10.1093/hmg/5.10.1679. [DOI] [PubMed] [Google Scholar]
- Olavesen M. G., Hampe J., Mirza M. M., Saiz R., Lewis C. M., Bridger S., Teare D., Easton D. F., Herrmann T., Scott G. Analysis of single-nucleotide polymorphisms in the interleukin-4 receptor gene for association with inflammatory bowel disease. Immunogenetics. 2000 Jan;51(1):1–7. doi: 10.1007/s002510050001. [DOI] [PubMed] [Google Scholar]
- Orholm M., Munkholm P., Langholz E., Nielsen O. H., Sørensen T. I., Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991 Jan 10;324(2):84–88. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- Podolsky D. K. Inflammatory bowel disease (1) N Engl J Med. 1991 Sep 26;325(13):928–937. doi: 10.1056/NEJM199109263251306. [DOI] [PubMed] [Google Scholar]
- Rioux J. D., Daly M. J., Green T., Stone V., Lander E. S., Hudson T. J., Steinhart A. H., Bull S., Cohen Z., Greenberg G. Absence of linkage between inflammatory bowel disease and selected loci on chromosomes 3, 7, 12, and 16. Gastroenterology. 1998 Nov;115(5):1062–1065. doi: 10.1016/s0016-5085(98)70074-5. [DOI] [PubMed] [Google Scholar]
- Roth M. P., Petersen G. M., McElree C., Vadheim C. M., Panish J. F., Rotter J. I. Familial empiric risk estimates of inflammatory bowel disease in Ashkenazi Jews. Gastroenterology. 1989 Apr;96(4):1016–1020. doi: 10.1016/0016-5085(89)91618-1. [DOI] [PubMed] [Google Scholar]
- Satsangi J., Parkes M., Louis E., Hashimoto L., Kato N., Welsh K., Terwilliger J. D., Lathrop G. M., Bell J. I., Jewell D. P. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996 Oct;14(2):199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- Slager S. L., Huang J., Vieland V. J. Effect of allelic heterogeneity on the power of the transmission disequilibrium test. Genet Epidemiol. 2000 Feb;18(2):143–156. doi: 10.1002/(SICI)1098-2272(200002)18:2<143::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Smith M. W., Dean M., Carrington M., Winkler C., Huttley G. A., Lomb D. A., Goedert J. J., O'Brien T. R., Jacobson L. P., Kaslow R. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997 Aug 15;277(5328):959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- Spielman R. S., McGinnis R. E., Ewens W. J. The transmission/disequilibrium test detects cosegregation and linkage. Am J Hum Genet. 1994 Mar;54(3):559–563. [PMC free article] [PubMed] [Google Scholar]
- Terwilliger J. D. A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet. 1995 Mar;56(3):777–787. [PMC free article] [PubMed] [Google Scholar]
- Thompson N. P., Driscoll R., Pounder R. E., Wakefield A. J. Genetics versus environment in inflammatory bowel disease: results of a British twin study. BMJ. 1996 Jan 13;312(7023):95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tysk C., Lindberg E., Järnerot G., Flodérus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988 Jul;29(7):990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A. F., Carothers A. D., Pirastu M. Population choice in mapping genes for complex diseases. Nat Genet. 1999 Dec;23(4):397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- Ye X., Mehlen P., Rabizadeh S., VanArsdale T., Zhang H., Shin H., Wang J. J., Leo E., Zapata J., Hauser C. A. TRAF family proteins interact with the common neurotrophin receptor and modulate apoptosis induction. J Biol Chem. 1999 Oct 15;274(42):30202–30208. doi: 10.1074/jbc.274.42.30202. [DOI] [PubMed] [Google Scholar]