Abstract
BACKGROUND—Gluten sensitivity is a common multifactorial disorder, manifested in the small intestine or on the skin as typical coeliac disease or dermatitis herpetiformis, respectively. The only established genetic risk factor is HLA DQ2. AIMS—We tested genetic linkage of previously reported chromosomal loci 5q and 11q in Finnish families with gluten sensitivity. We also tested if genetic linkage to candidate loci on 5q, 11q, 2q33, and HLA DQ differed with respect to clinical manifestations or sex. SUBJECTS—We studied 102 Finnish families with affected sibpairs. For heterogeneity analysis, families were divided into subgroups according to sex and the presence of dermatitis herpetiformis, the skin manifestation of gluten sensitivity. METHODS—Non-parametric linkage between microsatellite markers and disease was tested. Linkage heterogeneity between subgroups was tested using the M test. The transmission/disequilibrium test and association analysis were performed. RESULTS—Evidence of linkage to 11q (MLS 1.37), but not to 5q, was found in the entire dataset of 102 families. Heterogeneity between subgroups was suggested: families with only the intestinal disease showed linkage mainly to 2q33 whereas families with dermatitis herpetiformis showed linkage to 11q and 5q, but not to 2q33. Linkage in all three non-HLA loci was strongest in families with predominantly male patients. HLA DQ2 conferred much stronger susceptibility to females than males. CONCLUSIONS—Independent evidence for the suggested genetic linkage between 11q and gluten sensitivity was obtained. The possible linkage heterogeneity suggests genetic differences between intestinal and skin manifestations, and the gender dependent effect of HLA DQ2. Keywords: gluten sensitive enteropathy; coeliac disease; dermatitis herpetiformis; Finnish population; linkage analysis
Full Text
The Full Text of this article is available as a PDF (149.0 KB).
Figure 1 .
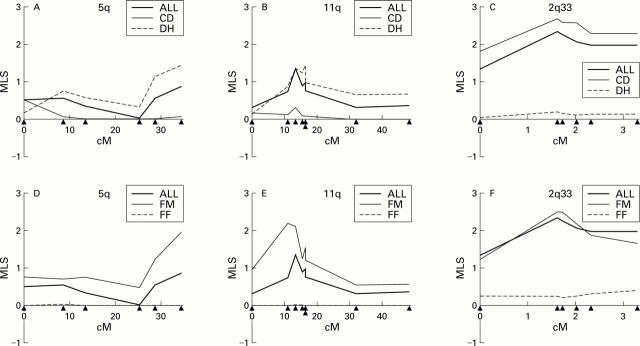
Maximum likelihood scores (MLS) in candidate regions 5q, 11q, and CD28/CTLA4 on 2q33 in the entire study group of 102 families and in the subgroups. Group ALL (all 102 families) was divided into group DH (dermatitis herpetiformis) and group CD (absence of dermatitis herpetiformis) (A-C) or group FM (male patients among affected siblings) and group FF (absence of male patients among affected siblings) (D-F). Marker positions are shown as triangles, from left to right: at 5q, D5S410, D5S422, D5S2032, D5S425, D5S2069, and D5S2111; at 11q, D11S898, D11S4111, D11S4142, D11S976, D11S4171, CD3D, D11S934, and D11S910; and at 2q33, D2S2392, D2S116, D2S2214, CTLA4(AT)n, D2S2189, and D2S2237.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brett P. M., Yiannakou J. Y., Morris M. A., Bronson S. R., Mathew C., Curtis D., Ciclitira P. J. A pedigree-based linkage study of coeliac disease: failure to replicate previous positive findings. Ann Hum Genet. 1998 Jan;62(Pt 1):25–32. doi: 10.1046/j.1469-1809.1998.6210025.x. [DOI] [PubMed] [Google Scholar]
- Djilali-Saiah I., Schmitz J., Harfouch-Hammoud E., Mougenot J. F., Bach J. F., Caillat-Zucman S. CTLA-4 gene polymorphism is associated with predisposition to coeliac disease. Gut. 1998 Aug;43(2):187–189. doi: 10.1136/gut.43.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco L., Corazza G., Babron M. C., Clot F., Fulchignoni-Lataud M. C., Percopo S., Zavattari P., Bouguerra F., Dib C., Tosi R. Genome search in celiac disease. Am J Hum Genet. 1998 Mar;62(3):669–675. doi: 10.1086/301754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holopainen P., Arvas M., Sistonen P., Mustalahti K., Collin P., Mäki M., Partanen J. CD28/CTLA4 gene region on chromosome 2q33 confers genetic susceptibility to celiac disease. A linkage and family-based association study. Tissue Antigens. 1999 May;53(5):470–475. doi: 10.1034/j.1399-0039.1999.530503.x. [DOI] [PubMed] [Google Scholar]
- Houlston R. S., Tomlinson I. P., Ford D., Seal S., Marossy A. M., Ferguson A., Holmes G. K., Hosie K. B., Howdle P. D., Jewell D. P. Linkage analysis of candidate regions for coeliac disease genes. Hum Mol Genet. 1997 Aug;6(8):1335–1339. doi: 10.1093/hmg/6.8.1335. [DOI] [PubMed] [Google Scholar]
- Kruglyak L., Lander E. S. Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet. 1995 Aug;57(2):439–454. [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen S., Gschwend M., Rioux J. D., Daly M. J., Terwilliger J. D., Tienari P. J., Wikström J., Palo J., Stein L. D., Hudson T. J. Genomewide scan of multiple sclerosis in Finnish multiplex families. Am J Hum Genet. 1997 Dec;61(6):1379–1387. doi: 10.1086/301637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan G., Huang D., Tait B. D., Colman P. G., Harrison L. C. Markers on distal chromosome 2q linked to insulin-dependent diabetes mellitus. Science. 1996 Jun 21;272(5269):1811–1813. doi: 10.1126/science.272.5269.1811. [DOI] [PubMed] [Google Scholar]
- Mäki M., Collin P. Coeliac disease. Lancet. 1997 Jun 14;349(9067):1755–1759. doi: 10.1016/S0140-6736(96)70237-4. [DOI] [PubMed] [Google Scholar]
- Paterson A. D., Petronis A. Sex of affected sibpairs and genetic linkage to type 1 diabetes. Am J Med Genet. 1999 May 7;84(1):15–19. doi: 10.1002/(sici)1096-8628(19990507)84:1<15::aid-ajmg4>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Peltonen L., Jalanko A., Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet. 1999;8(10):1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- Petronzelli F., Bonamico M., Ferrante P., Grillo R., Mora B., Mariani P., Apollonio I., Gemme G., Mazzilli M. C. Genetic contribution of the HLA region to the familial clustering of coeliac disease. Ann Hum Genet. 1997 Jul;61(Pt 4):307–317. doi: 10.1046/j.1469-1809.1997.6140307.x. [DOI] [PubMed] [Google Scholar]
- Polvi A., Arranz E., Fernandez-Arquero M., Collin P., Mäki M., Sanz A., Calvo C., Maluenda C., Westman P., de la Concha E. G. HLA-DQ2-negative celiac disease in Finland and Spain. Hum Immunol. 1998 Mar;59(3):169–175. doi: 10.1016/s0198-8859(98)00008-1. [DOI] [PubMed] [Google Scholar]
- Reunala T., Kosnai I., Karpati S., Kuitunen P., Török E., Savilahti E. Dermatitis herpetiformis: jejunal findings and skin response to gluten free diet. Arch Dis Child. 1984 Jun;59(6):517–522. doi: 10.1136/adc.59.6.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid L. M., Thorsby E. HLA susceptibility genes in celiac disease: genetic mapping and role in pathogenesis. Gastroenterology. 1993 Sep;105(3):910–922. doi: 10.1016/0016-5085(93)90912-v. [DOI] [PubMed] [Google Scholar]
- Terwilliger J. D., Weiss K. M. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol. 1998 Dec;9(6):578–594. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]
- Vaidya B., Imrie H., Perros P., Young E. T., Kelly W. F., Carr D., Large D. M., Toft A. D., McCarthy M. I., Kendall-Taylor P. The cytotoxic T lymphocyte antigen-4 is a major Graves' disease locus. Hum Mol Genet. 1999 Jul;8(7):1195–1199. doi: 10.1093/hmg/8.7.1195. [DOI] [PubMed] [Google Scholar]
- Zhong F., McCombs C. C., Olson J. M., Elston R. C., Stevens F. M., McCarthy C. F., Michalski J. P. An autosomal screen for genes that predispose to celiac disease in the western counties of Ireland. Nat Genet. 1996 Nov;14(3):329–333. doi: 10.1038/ng1196-329. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A. Disease gene mapping in isolated human populations: the example of Finland. J Med Genet. 1993 Oct;30(10):857–865. doi: 10.1136/jmg.30.10.857. [DOI] [PMC free article] [PubMed] [Google Scholar]