Abstract
BACKGROUND AND AIMS—Chronic hepatitis C is a slowly progressive disease and eventually causes hepatocellular carcinoma in many patients. Although interferon (IFN) therapy has been used for viral eradication, its success rate is only about 30%. In patients in whom it has failed (non-responders), there are several patterns of serum alanine aminotransferase (ALT) values, and detection of serum HCV-RNA during and after IFN therapy and improved long term prognosis were reported in patients whose serum ALT values were normalised by IFN therapy even if HCV viraemia persisted. The present study sought to clarify the virological characteristics contributing to these differences. METHODS—Complete or partial length dominant sequences of hepatitis C virus genotype 1b (HCV-1b) were determined by direct sequencing. Firstly, the complete sequences of HCV-1b genomes were determined in six non-responders; three showed normalisation of serum ALT values during IFN-α therapy and the other three did not. Subsequently, the amino acid residues that were different in the two groups were further analysed retrospectively in another 82 patients. RESULTS—Comparison of the sequences suggested an association between amino acids 2154-2172 of HCV-1b and serum ALT normalisation. A retrospective analysis of 82 patients revealed that the number of amino acid substitutions in this region was the only statistically significant variable associated with ALT normalisation (odds ratio 31.0; 95% confidence interval 5.0-286) in multivariate analyses. CONCLUSIONS—A HCV genomic region that correlates with the ALT response to IFN therapy appears to be present in virologically IFN ineffective patients. Keywords: hepatitis C virus; alanine aminotransferase; biochemical responder; transient responder; NS5A protein
Full Text
The Full Text of this article is available as a PDF (171.9 KB).
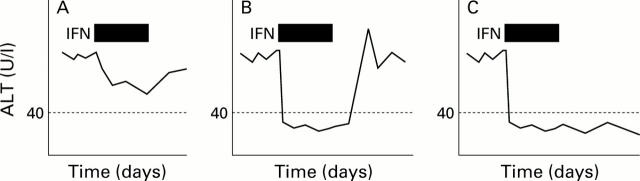
Figure 1 .
Schematic diagram of the typical clinical courses of serum alanine aminotransferase (ALT) values during and after interferon (IFN) therapy. (A) Patient without ALT normalisation (biochemical non-responder (bNR)); (B) patient with ALT normalisation and subsequent flare up (biochemical transient responder (bTR)); and (C) patient with ALT normalisation and sustained normal ALT values after IFN therapy (biochemical responder (BR)).
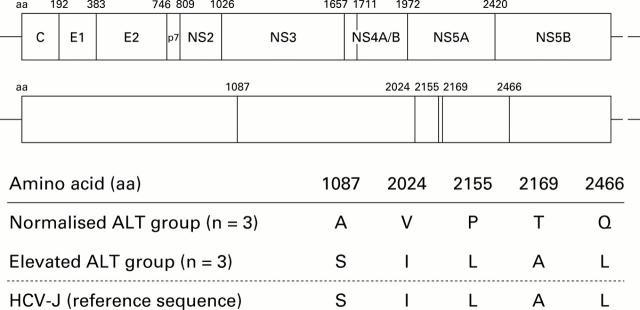
Figure 2 .
Codons for which the encoded amino acid are different between the two groups. The upper column shows the architecture of the hepatitis C virus (HCV) polyprotein and the lower column shows the distribution of the amino acid residues. Amino acid usages in each group and HCV-J are shown in the lowermost table. Amino acids are described in the single letter code.
Figure 3 .

Relationship between amino acid sequences in the alanine aminotransferase (ALT) response related element (ARE) in 82 patients. Amino acid residues from 2154 to 2172 are shown. Amino acid residues identical to those of HCV-J are shown as "-". Filled circles (•) indicates that serum ALT values normalised during interferon therapy. *Biochemical responders.
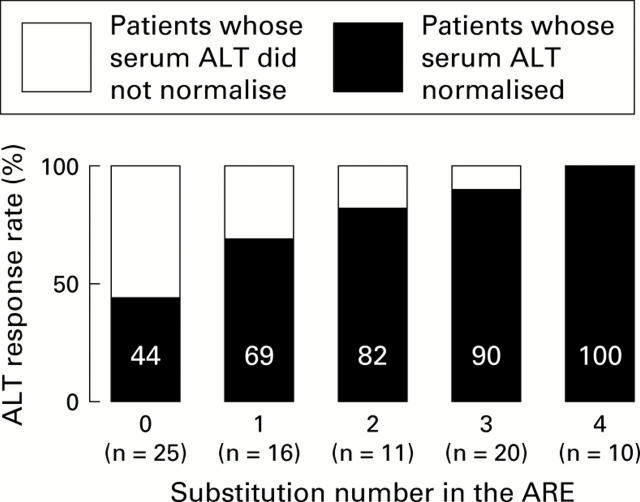
Figure 4 .
Relationship between substitution number in the alanine aminotransferase (ALT) response related element (ARE) and the proportion of patients with ALT normalisation. The number of patients in each group is shown in parentheses.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cammà C., Giunta M., Linea C., Pagliaro L. The effect of interferon on the liver in chronic hepatitis C: a quantitative evaluation of histology by meta-analysis. J Hepatol. 1997 Jun;26(6):1187–1199. doi: 10.1016/s0168-8278(97)80451-5. [DOI] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Desmet V. J., Gerber M., Hoofnagle J. H., Manns M., Scheuer P. J. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994 Jun;19(6):1513–1520. [PubMed] [Google Scholar]
- Duverlie G., Khorsi H., Castelain S., Jaillon O., Izopet J., Lunel F., Eb F., Penin F., Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998 Jun;79(Pt 6):1373–1381. doi: 10.1099/0022-1317-79-6-1373. [DOI] [PubMed] [Google Scholar]
- Enomoto N., Sakuma I., Asahina Y., Kurosaki M., Murakami T., Yamamoto C., Izumi N., Marumo F., Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995 Jul;96(1):224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto N., Sakuma I., Asahina Y., Kurosaki M., Murakami T., Yamamoto C., Ogura Y., Izumi N., Marumo F., Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996 Jan 11;334(2):77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- Fukuma T., Enomoto N., Marumo F., Sato C. Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein. Hepatology. 1998 Oct;28(4):1147–1153. doi: 10.1002/hep.510280433. [DOI] [PubMed] [Google Scholar]
- Hoofnagle J. H., di Bisceglie A. M. The treatment of chronic viral hepatitis. N Engl J Med. 1997 Jan 30;336(5):347–356. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Saitoh S., Arase Y., Chayama K., Suzuki Y., Kobayashi M., Tsubota A., Nakamura I., Murashima N., Kumada H. Effect of interferon therapy on hepatocellular carcinogenesis in patients with chronic hepatitis type C: A long-term observation study of 1,643 patients using statistical bias correction with proportional hazard analysis. Hepatology. 1999 Apr;29(4):1124–1130. doi: 10.1002/hep.510290439. [DOI] [PubMed] [Google Scholar]
- Kasahara A., Hayashi N., Mochizuki K., Takayanagi M., Yoshioka K., Kakumu S., Iijima A., Urushihara A., Kiyosawa K., Okuda M. Risk factors for hepatocellular carcinoma and its incidence after interferon treatment in patients with chronic hepatitis C. Osaka Liver Disease Study Group. Hepatology. 1998 May;27(5):1394–1402. doi: 10.1002/hep.510270529. [DOI] [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Lan K. H., Ono-Nita S. K., Shiratori Y., Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997 Nov;71(11):8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa K., Sodeyama T., Tanaka E., Gibo Y., Yoshizawa K., Nakano Y., Furuta S., Akahane Y., Nishioka K., Purcell R. H. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990 Oct;12(4 Pt 1):671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- Kuzushita N., Hayashi N., Kanto T., Takehara T., Tatsumi T., Katayama K., Ohkawa K., Ito A., Kasahara A., Moribe T. Involvement of transporter associated with antigen processing 2 (TAP2) gene polymorphisms in hepatitis C virus infection. Gastroenterology. 1999 May;116(5):1149–1154. doi: 10.1016/s0016-5085(99)70018-1. [DOI] [PubMed] [Google Scholar]
- Lau J. Y., Davis G. L., Kniffen J., Qian K. P., Urdea M. S., Chan C. S., Mizokami M., Neuwald P. D., Wilber J. C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993 Jun 12;341(8859):1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- Major M. E., Feinstone S. M. The molecular virology of hepatitis C. Hepatology. 1997 Jun;25(6):1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- Markland W., Petrillo R. A., Fitzgibbon M., Fox T., McCarrick R., McQuaid T., Fulghum J. R., Chen W., Fleming M. A., Thomson J. A. Purification and characterization of the NS3 serine protease domain of hepatitis C virus expressed in Saccharomyces cerevisiae. J Gen Virol. 1997 Jan;78(Pt 1):39–43. doi: 10.1099/0022-1317-78-1-39. [DOI] [PubMed] [Google Scholar]
- McMinn P. C. The molecular basis of virulence of the encephalitogenic flaviviruses. J Gen Virol. 1997 Nov;78(Pt 11):2711–2722. doi: 10.1099/0022-1317-78-11-2711. [DOI] [PubMed] [Google Scholar]
- Nagayama K., Kurosaki M., Enomoto N., Maekawa S. Y., Miyasaka Y., Tazawa J., Izumi N., Marumo F., Sato C. Time-related changes in full-length hepatitis C virus sequences and hepatitis activity. Virology. 1999 Oct 10;263(1):244–253. doi: 10.1006/viro.1999.9924. [DOI] [PubMed] [Google Scholar]
- Nakano I., Fukuda Y., Katano Y., Nakano S., Kumada T., Hayakawa T. Why is the interferon sensitivity-determining region (ISDR) system useful in Japan? J Hepatol. 1999 Jun;30(6):1014–1022. doi: 10.1016/s0168-8278(99)80254-2. [DOI] [PubMed] [Google Scholar]
- Nishiguchi S., Kuroki T., Nakatani S., Morimoto H., Takeda T., Nakajima S., Shiomi S., Seki S., Kobayashi K., Otani S. Randomised trial of effects of interferon-alpha on incidence of hepatocellular carcinoma in chronic active hepatitis C with cirrhosis. Lancet. 1995 Oct 21;346(8982):1051–1055. doi: 10.1016/s0140-6736(95)91739-x. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Sugiyama Y., Okada S., Kurai K., Akahane Y., Sugai Y., Tanaka T., Sato K., Tsuda F., Miyakawa Y. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992 Mar;73(Pt 3):673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- Peano G., Menardi G., Ponzetto A., Fenoglio L. M. HLA-DR5 antigen. A genetic factor influencing the outcome of hepatitis C virus infection? Arch Intern Med. 1994 Dec 12;154(23):2733–2736. doi: 10.1001/archinte.1994.00420230126015. [DOI] [PubMed] [Google Scholar]
- Poynard T., Leroy V., Cohard M., Thevenot T., Mathurin P., Opolon P., Zarski J. P. Meta-analysis of interferon randomized trials in the treatment of viral hepatitis C: effects of dose and duration. Hepatology. 1996 Oct;24(4):778–789. doi: 10.1002/hep.510240405. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Alberti A., Alter H. J., Bonino F., Bradley D. W., Brechot C., Brouwer J. T., Chan S. W., Chayama K., Chen D. S. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994 May;19(5):1321–1324. [PubMed] [Google Scholar]
- Tanaka T., Kato N., Cho M. J., Sugiyama K., Shimotohno K. Structure of the 3' terminus of the hepatitis C virus genome. J Virol. 1996 May;70(5):3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto A., Ide Y., Arima N., Sasaguri Y., Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun. 1997 Jul 18;236(2):360–364. doi: 10.1006/bbrc.1997.6967. [DOI] [PubMed] [Google Scholar]