Abstract
BACKGROUND—In liver regeneration after portal branch ligation we previously showed that early cellular changes are observed in both the proliferating and atrophying liver lobes. They are therefore not indicative of future proliferative response. In this study we attempted to define precisely, in the same model, the time at which the cellular processes diverge between the lobes by measuring various parameters associated with cellular proliferation. We also investigated the possible role of inhibitors of cell proliferation in the absence of progression towards the S phase in the atrophying lobes. AIMS—Expression of p53, c-Ha-ras, cyclin E, cyclin dependent kinase (Cdk2), transforming growth factor (TGF)-β, and interleukin (IL)-1α and IL-1β were assessed in relation to their potential role in proliferating and atrophying cellular phenomenons. METHODS—Immunohistochemistry, northern blotting, western blotting, and reverse transcription-polymerase chain reaction were performed, mainly at time points corresponding to mid-G1/S phase progression (8-24 hours after surgery). RESULTS—The common and thus most likely non-specific response was still evident 5-8 hours after surgery and included an increase in IL-1 mRNA as well as p53 and cyclin E proteins. From 12 hours onwards, p53, c-Ha-ras, cyclin E, and Cdk2 were selectively induced in proliferating lobes whereas IL-1β was predominantly activated in atrophying lobes. No changes in TGF-β or IL-1α expression were observed at the same time points in any of the liver lobes. CONCLUSIONS—The initial response to portal branch ligation and thus probably to partial hepatectomy seems to be non-specific for at least eight hours. Thereafter, p53, c-Ha-ras, cyclin E, and Cdk2 seem to drive cellular proliferation while IL-1β is associated with cellular atrophy. In contrast, TGF-β and IL-1α do not seem to play a role in determining the commitment of cells towards atrophy or proliferation. Keywords: portal branch ligation; liver regeneration; delayed early proto-oncogenes; cytokines; cyclin dependent kinase; rat
Full Text
The Full Text of this article is available as a PDF (278.2 KB).
Figure 1 .
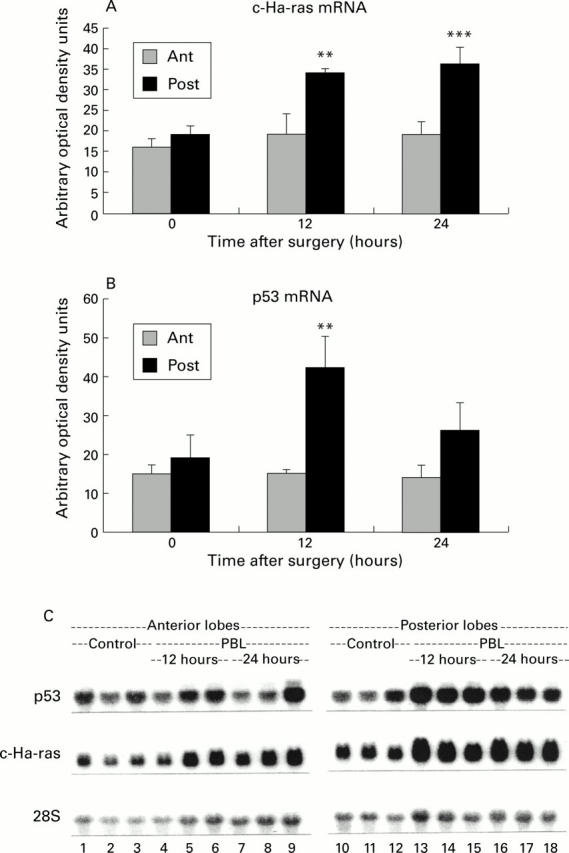
Northern blot analysis of p53 and c-Ha-ras mRNA. (A) Quantification of c-Ha-ras mRNA by densitometric analysis of the blots. The adjusted values did not reveal any significant rise in c-Ha-ras mRNA in the anterior lobes. In the posterior lobes, a significant increase in c-Ha-ras level was noticed from 12 hours after portal branch ligation (PBL). **p<0.01, ***p<0.001. (B) Quantification of p53 mRNA by densitometric analysis of the blots. After adjustment for the respective 28S ribosomal signals, no elevation above baseline levels was observed in atrophying (Ant) lobes whereas p53 mRNA was significantly upregulated (**p<0.01) in regenerating (Post) lobes at 12 hours after PBL. (C) Northern blots of p53, c-Ha-ras, and 28S mRNAs extracted from control animals (lanes 1-3 and 10-12) as well as from the anterior and posterior lobes of PBL animals (lanes 4-9 and 13-18, respectively). Total liver mRNA (20 µg) was separated by electrophoresis on agarose gels, hybridised with their respective 32P labelled cDNAs, and blotted. Signals were analysed by densitometry and adjusted for their 28S signals.
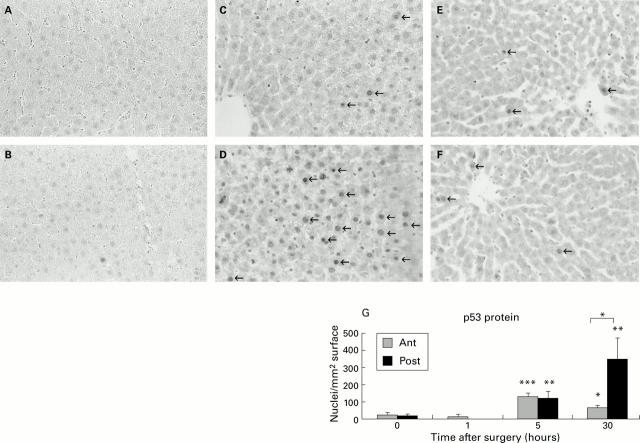
Figure 2 .
Immunohistochemistry of p53 protein expression after portal branch ligation (PBL). (A-F) Histology and p53 nuclear staining. Slides were stained immunohistochemically with a polyclonal anti-p53 antibody. No staining is observed in the anterior and posterior lobes of control animals (A, B). At five hours, nuclear staining (indicated by arrows) is slightly enhanced in both parts of the liver (E, F). At 30 hours, no further increase in p53 staining is noticed in the anterior lobes (C) whereas marked upregulation in positive staining nuclei is seen in the posterior lobes (D). (G) Quantification of p53 labelled nuclei. The labelled nuclei were counted in the anterior (Ant) and posterior (Post) lobes of control (0 hours) and PBL animals at one, five, and 30 hours. The final result was adjusted for the surface (mm2) of the section. A similar and significant increase in p53 positive nuclei was noticed in the regenerating (Post) and atrophying (Ant) lobes at five hours after PBL (**p<0.01 and ***p<0.001, respectively) whereas at 30 hours a further rise in p53 positive nuclei was only shown in the regenerating lobes. At this time point a significant difference between the lobes was observed (*p<0.05).
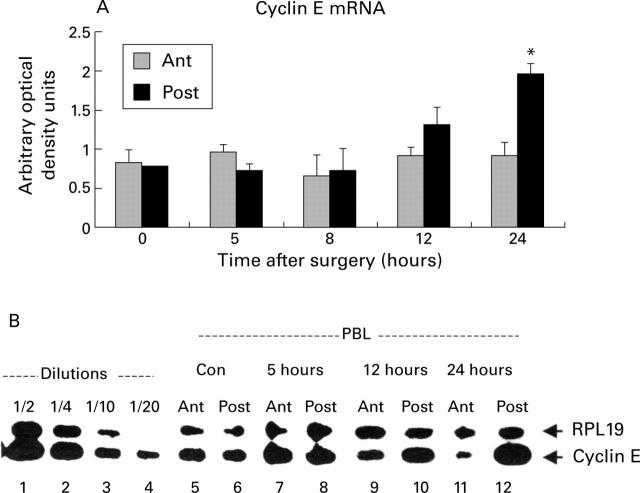
Figure 3 .
Expression of cyclin E mRNA in the atrophying and regenerating lobes after portal branch ligation (PBL). (A) Quantification of cyclin E mRNA by densitometric analysis. Total liver RNA obtained from the atrophying (Ant) and regenerating (Post) lobes was subjected to reverse transcription-polymerase chain reaction (RT-PCR), as described in materials and methods. The results were adjusted for their respective RPL19 levels and then compared with a standard dilution curve obtained by amplification of a cyclin E mRNA sample in the same PCR procedure. A significant induction of cyclin E mRNA was only observed in the regenerating lobes at 24 hours after PBL (*p<0.05). (B) RT-PCR of cyclin E mRNA. Representation of RT-PCR of RPL19 and cyclin E mRNA in control animals (lanes 5 and 6) and in the atrophying (Ant) and regenerating (Post) lobes at various time points after PBL (lanes 7-12). Total liver RNA was subjected to RT-PCR as described in materials and methods. The results were adjusted for their respective RPL19 levels and compared with a standard dilution curve obtained by amplification of liver mRNA from lipopolysaccharide treated rats in the same PCR procedure (lanes 1-4).
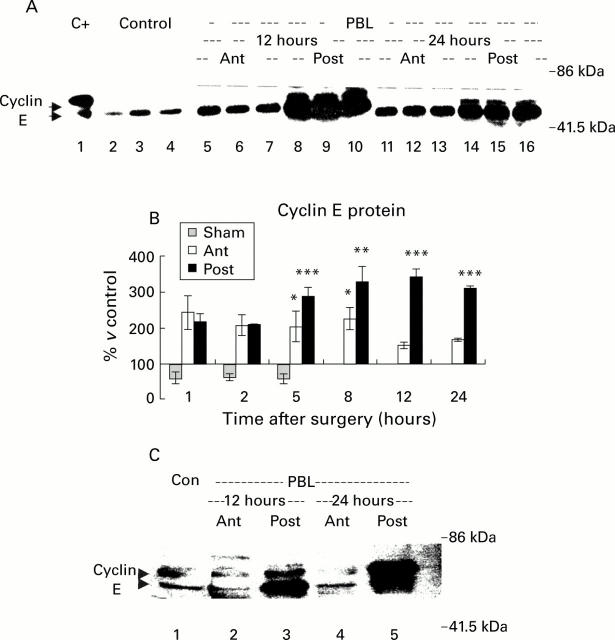
Figure 4 .
Western blot of cyclin E protein. (A) Cytoplasmic proteins (40 µg) were resolved using 8% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and blotted with an anti-cyclin E polyclonal antibody. Samples from control animals (0 hours) and from the atrophying (Ant) and regenerating (Post) lobes of animals at various times after portal branch ligation (PBL) were analysed. The antibody recognised two bands of approximately 52 and 54 kDa in control and PBL animals. A representative blot of three control (lanes 2-4) and three PBL cytosol extracts at 12 hours (lanes 5-10) and 24 hours (lanes 11-16) after surgery is shown. KNRK nuclear extracts were used as a positive control (C+, lane 1). (B) Quantification of cyclin E protein in the cytoplasm by densitometry. Cyclin E protein was rapidly upregulated in both lobes within the first hours after PBL. A further significant increase was observed in the regenerating lobes between 8 and 24 hours after PBL (**p<0.01, ***p<0.001, respectively). In contrast, cyclin E protein expression decreased in the atrophying lobes during the same time without, however, reaching control levels. (C) Nuclear proteins (40 µg) were subjected to western blot analysis. Very low amounts of cyclin E protein were detected in the nuclear fraction of control livers (Con). A faint band of approximately 52 kDa was found in nuclear fractions of both lobes at all investigated time points which increased selectively in the regenerating lobes (Post) from 12 hours onwards (lane 3). A slower migrating approximately 54 kD band, initially weakly present in both parts, was strongly upregulated in the nuclear fractions from the regenerating (Post) lobes at 24 hours (lane 5).
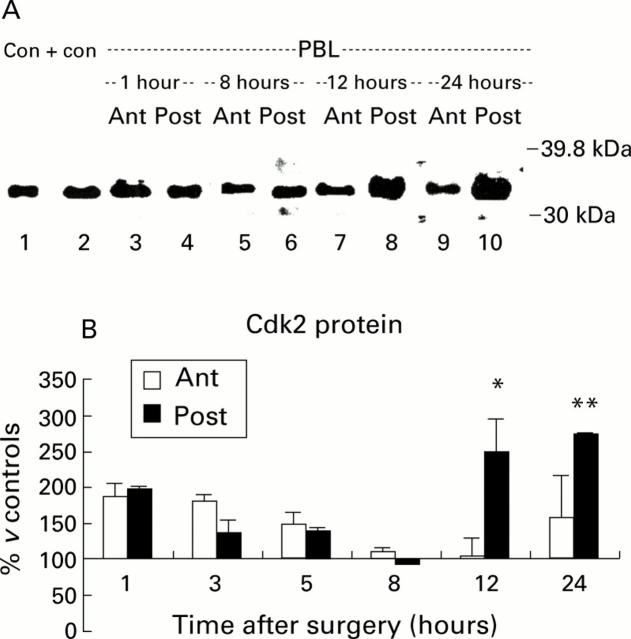
Figure 5 .
Western blot of cyclin dependent kinase (Cdk2) protein in the cytoplasm. (A) Cytoplasmic proteins (30 µg) were resolved using 8% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and blotted with an anti-Cdk2 polyclonal antibody. Samples from control animals (0 hours) and from the atrophying (Ant) and regenerating (Post) lobes of animals at various times after portal branch ligation (PBL) were analysed. An approximately 34 kDa band was found in control livers (Con, lane 2) and in both lobes after PBL. Increased expression was observed in the regenerating (Post) lobes mainly at 12 and 24 hours after PBL (lanes 8 and 10, respectively). KNKR nuclear extracts were used as a positive control (Con+, lane 1). (B) Quantification of Cdk2 protein in the cytoplasm by densitometry. Cdk2 was similarly increased in both lobes during the very first hours after PBL but returned to control levels at eight hours. Thereafter, Cdk2 increased again significantly (*p<0.05;**p<0.01) only in the regenerating (Post) lobes until 24 hours after PBL.
Figure 6 .
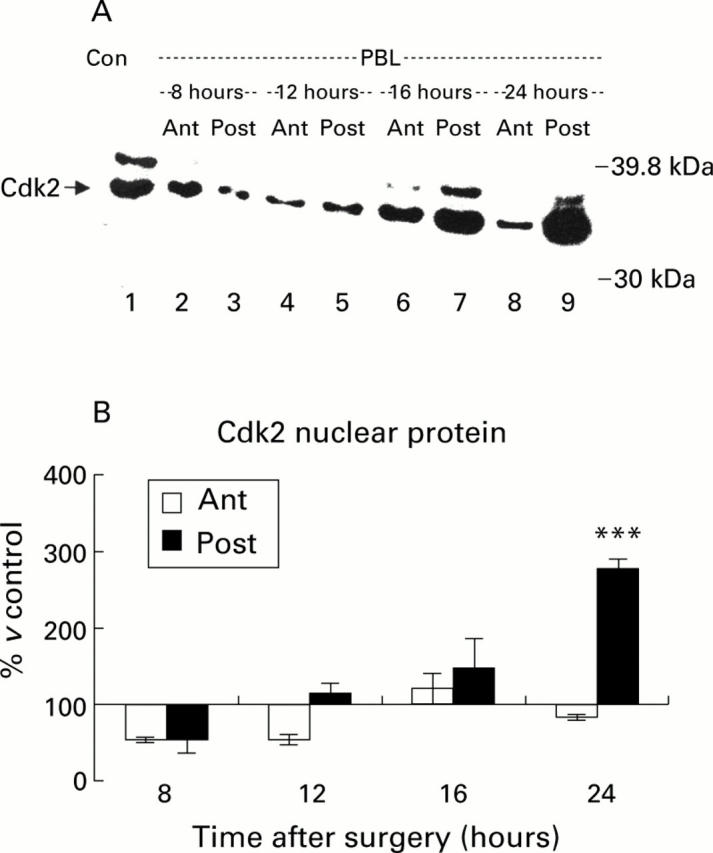
Western blot of cyclin dependent kinase (Cdk2) protein in the nuclear fraction. (A) Cytoplasmic proteins (30 µg) were resolved using 8% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and blotted with an anti-Cdk2 polyclonal antibody. Samples from control animals (Con) and from the atrophying (Ant) and regenerating (Post) lobes of animals at various times after portal branch ligation (PBL) were analysed. Low levels of Cdk2 were found in control livers (Con, lane 1) and in the atrophying (Ant) lobes after PBL. Increased expression was observed in the regenerating (Post) lobes from 16 hours onwards, reaching a peak at 24 hours after PBL (lanes 7 and 9, respectively). (B) Quantification of Cdk2 protein in the nuclear fraction by densitometry. No significant increase in Cdk2 expression compared with control levels was seen in the atrophying (Ant) lobes after PBL. In the regenerating (Post) lobes however Cdk2 levels started to rise from 12 hours after PBL and reached a significant peak (***p<0.001) at 24 hours after PBL.
Figure 7 .
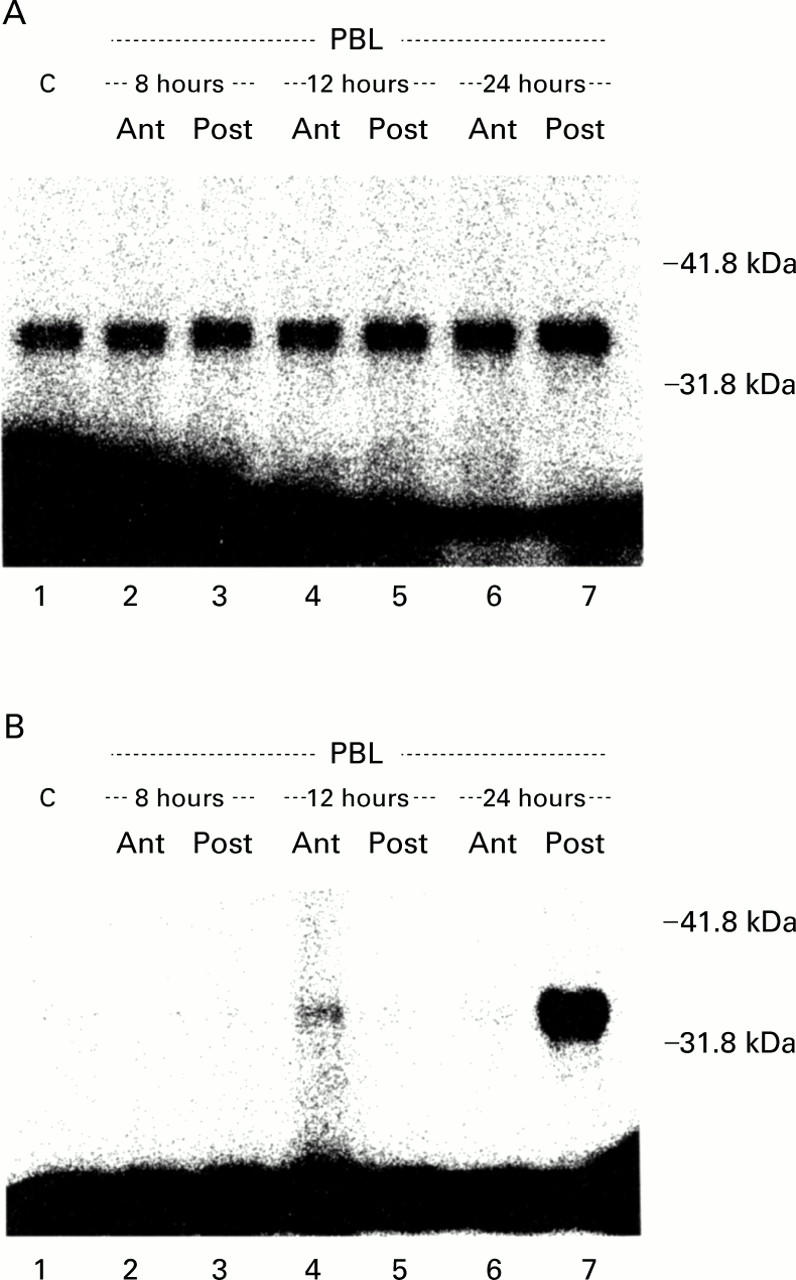
Cyclin dependent kinase (Cdk2) associated activity in cytoplasmic and nuclear fractions. Cdk2 protein was immunoprecipitated and a histone H1 kinase assay was performed followed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. (A) Low constitutive activity was detected in the cytosol of control livers (C) and in the atrophying (Ant) and regenerating (Post) lobes after portal branch ligation (PBL) (lanes 2-7) which did not significantly vary during the experiment. (B) In contrast, no Cdk2 activity was observed in the nuclear fractions of control (C, lane 1) and PBL livers (lanes 2-6) until 24 hours. At 24 hours after PBL a sharp increase in Cdk2 activity was selectively found in the regenerating lobes (lane 7).
Figure 8 .
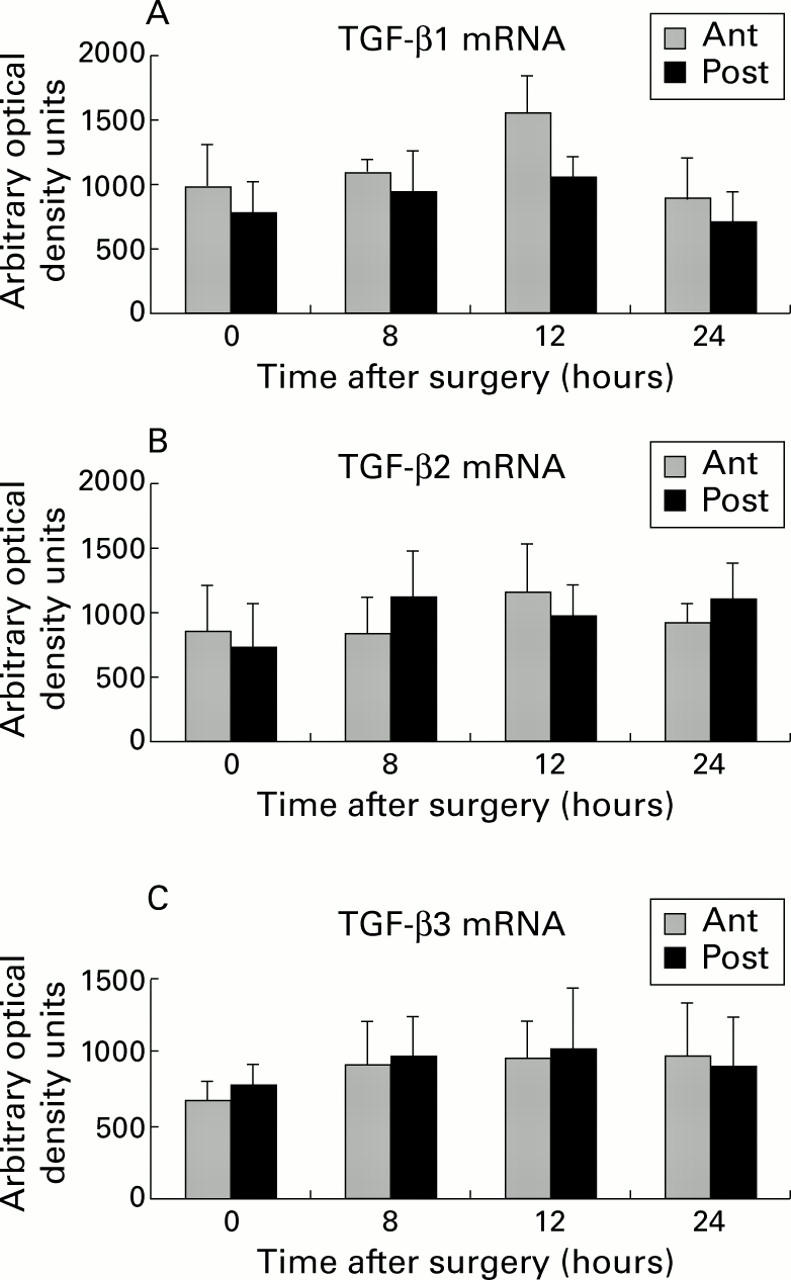
Quantification of transforming growth factor (TGF)-β1, -β2, and -β3 mRNA by densitometric analysis. Total liver RNA was subjected to reverse transcription-polymerase chain reaction, as described in materials and methods using TGF-β1 (A), TGF-β2 (B), and TGF-β3 (C) specific primers. The results were adjusted for their respective RPL19 levels and compared with a standard dilution curve obtained by amplification of liver mRNA from lipopolysaccharide treated rats in the same polymerase chain reaction procedure. Compared with control levels (0 hours), no significant differences in TGF-β mRNA levels were noted at any of the investigated time points after portal branch ligation (eight, 12, or 24 hours).
Figure 9 .
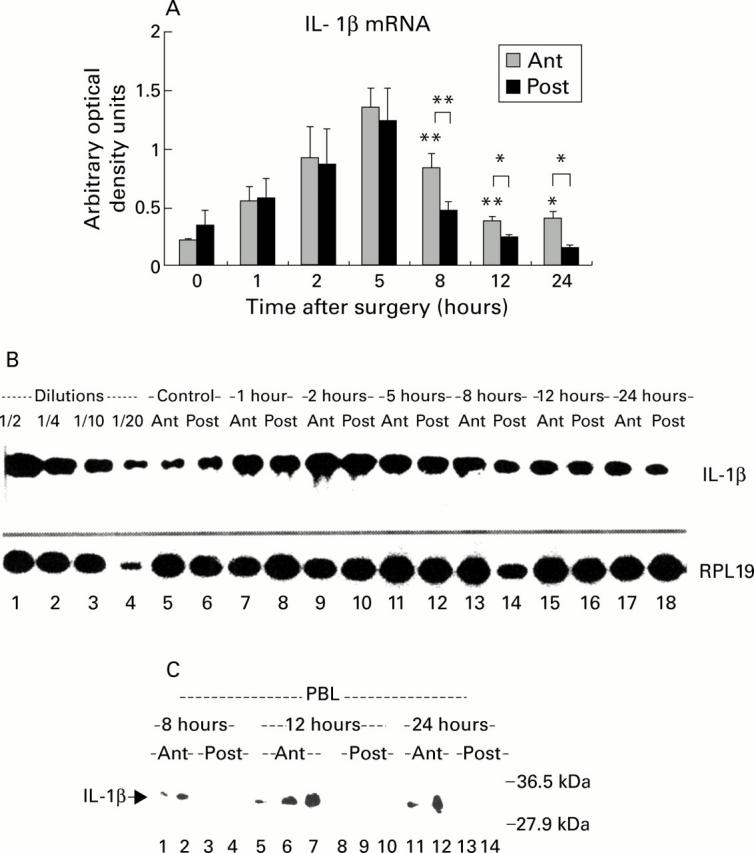
Expression of interleukin (IL)-1β in atrophying and regenerating lobes after portal branch ligation (PBL). (A) Quantification of IL-1β mRNA by densitometric analysis. IL-1β mRNA was significantly induced in both lobes during the first hours (1-5) after PBL. From eight hours after PBL, IL-1β mRNA returned to control levels in the regenerating lobes (Post) whereas significant activation of IL-1 persisted in the atrophying lobes (Ant) (*p<0.05, **p<0.01). (B) Reverse transcription-polymerase chain reaction (RT-PCR) of IL-1β mRNA. Representation of a RT-PCR of RPL19 and IL-1β mRNA in control animals (lanes 5-6) and in the atrophying (Ant) and regenerating (Post) lobes at various time points after portal branch ligation (lanes 7-18). Total liver RNA was subjected to RT-PCR, as described in materials and methods. The results were adjusted for their respective RPL19 levels and compared with a standard dilution curve obtained by amplification of liver mRNA from lipopolysaccharide treated rats in the same PCR procedure (lanes 1-4). (C) Western blot of IL-1β protein in liver homogenates. At high protein load (up to 300 µg), an approximately 31 kDa band (IL-1β precursor) was observed in the atrophying lobes at eight (lanes 1, 2), 12 (lanes 5-7), and 24 hours (lanes 11, 12) after PBL. No signal was detected in the proliferating lobes at the same time points (lanes 3, 4, 8-10, 13, 14, respectively).
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aktas H., Cai H., Cooper G. M. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol Cell Biol. 1997 Jul;17(7):3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht J. H., Rieland B. M., Nelsen C. J., Ahonen C. L. Regulation of G(1) cyclin-dependent kinases in the liver: role of nuclear localization and p27 sequestration. Am J Physiol. 1999 Dec;277(6 Pt 1):G1207–G1216. doi: 10.1152/ajpgi.1999.277.6.G1207. [DOI] [PubMed] [Google Scholar]
- Attardi L. D., Lowe S. W., Brugarolas J., Jacks T. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J. 1996 Jul 15;15(14):3693–3701. [PMC free article] [PubMed] [Google Scholar]
- Attisano L., Wrana J. L., López-Casillas F., Massagué J. TGF-beta receptors and actions. Biochim Biophys Acta. 1994 May 26;1222(1):71–80. doi: 10.1016/0167-4889(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Baker S. J., Markowitz S., Fearon E. R., Willson J. K., Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990 Aug 24;249(4971):912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- Bellamy C. O., Clarke A. R., Wyllie A. H., Harrison D. J. p53 Deficiency in liver reduces local control of survival and proliferation, but does not affect apoptosis after DNA damage. FASEB J. 1997 Jun;11(7):591–599. doi: 10.1096/fasebj.11.7.9212083. [DOI] [PubMed] [Google Scholar]
- Beyer H. S., Theologides A. Tumor necrosis factor-alpha is a direct hepatocyte mitogen in the rat. Biochem Mol Biol Int. 1993 Jan;29(1):1–4. [PubMed] [Google Scholar]
- Biesiada E., Chorazy M. Expression of "cell-cycle-dependent" genes in regenerating rat liver. Cell Biol Int Rep. 1988 Jun;12(6):483–492. doi: 10.1016/0309-1651(88)90140-3. [DOI] [PubMed] [Google Scholar]
- Bissell D. M., Wang S. S., Jarnagin W. R., Roll F. J. Cell-specific expression of transforming growth factor-beta in rat liver. Evidence for autocrine regulation of hepatocyte proliferation. J Clin Invest. 1995 Jul;96(1):447–455. doi: 10.1172/JCI118055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boermeester M. A., Straatsburg I. H., Houdijk A. P., Meyer C., Frederiks W. M., Wesdorp R. I., van Noorden C. J., van Leeuwen P. A. Endotoxin and interleukin-1 related hepatic inflammatory response promotes liver failure after partial hepatectomy. Hepatology. 1995 Nov;22(5):1499–1506. [PubMed] [Google Scholar]
- Boulton R. A., Alison M. R., Golding M., Selden C., Hodgson H. J. Augmentation of the early phase of liver regeneration after 70% partial hepatectomy in rats following selective Kupffer cell depletion. J Hepatol. 1998 Aug;29(2):271–280. doi: 10.1016/s0168-8278(98)80013-5. [DOI] [PubMed] [Google Scholar]
- Boulton R., Woodman A., Calnan D., Selden C., Tam F., Hodgson H. Nonparenchymal cells from regenerating rat liver generate interleukin-1alpha and -1beta: a mechanism of negative regulation of hepatocyte proliferation. Hepatology. 1997 Jul;26(1):49–58. doi: 10.1053/jhep.1997.v26.pm0009214451. [DOI] [PubMed] [Google Scholar]
- Braun L., Mead J. E., Panzica M., Mikumo R., Bell G. I., Fausto N. Transforming growth factor beta mRNA increases during liver regeneration: a possible paracrine mechanism of growth regulation. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1539–1543. doi: 10.1073/pnas.85.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L., Elledge S. J., Harper J. W. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr Biol. 1998 Jan 1;8(1):65–68. doi: 10.1016/s0960-9822(98)70021-1. [DOI] [PubMed] [Google Scholar]
- Danova M., Giordano M., Mazzini G., Riccardi A. Expression of p53 protein during the cell cycle measured by flow cytometry in human leukemia. Leuk Res. 1990;14(5):417–422. doi: 10.1016/0145-2126(90)90027-7. [DOI] [PubMed] [Google Scholar]
- Date M., Matsuzaki K., Matsushita M., Sakitani K., Shibano K., Okajima A., Yamamoto C., Ogata N., Okumura T., Seki T. Differential expression of transforming growth factor-beta and its receptors in hepatocytes and nonparenchymal cells of rat liver after CCl4 administration. J Hepatol. 1998 Apr;28(4):572–581. doi: 10.1016/s0168-8278(98)80280-8. [DOI] [PubMed] [Google Scholar]
- Deppert W., Buschhausen-Denker G., Patschinsky T., Steinmeyer K. Cell cycle control of p53 in normal (3T3) and chemically transformed (Meth A) mouse cells. II. Requirement for cell cycle progression. Oncogene. 1990 Nov;5(11):1701–1706. [PubMed] [Google Scholar]
- Dobrowolski S., Harter M., Stacey D. W. Cellular ras activity is required for passage through multiple points of the G0/G1 phase in BALB/c 3T3 cells. Mol Cell Biol. 1994 Aug;14(8):5441–5449. doi: 10.1128/mcb.14.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R. J., Brook A., Dyson N., O'Farrell P. H. E2F-induced S phase requires cyclin E. Genes Dev. 1996 Oct 1;10(19):2505–2513. doi: 10.1101/gad.10.19.2505. [DOI] [PubMed] [Google Scholar]
- Ehrenfried J. A., Ko T. C., Thompson E. A., Evers B. M. Cell cycle-mediated regulation of hepatic regeneration. Surgery. 1997 Nov;122(5):927–935. doi: 10.1016/s0039-6060(97)90334-2. [DOI] [PubMed] [Google Scholar]
- Fan G., Xu R., Wessendorf M. W., Ma X., Kren B. T., Steer C. J. Modulation of retinoblastoma and retinoblastoma-related proteins in regenerating rat liver and primary hepatocytes. Cell Growth Differ. 1995 Nov;6(11):1463–1476. [PubMed] [Google Scholar]
- Fausto N., Laird A. D., Webber E. M. Liver regeneration. 2. Role of growth factors and cytokines in hepatic regeneration. FASEB J. 1995 Dec;9(15):1527–1536. doi: 10.1096/fasebj.9.15.8529831. [DOI] [PubMed] [Google Scholar]
- Fausto N., Mead J. E. Regulation of liver growth: protooncogenes and transforming growth factors. Lab Invest. 1989 Jan;60(1):4–13. [PubMed] [Google Scholar]
- Fausto N., Mead J. E. Role of protooncogenes and transforming growth factors in normal and neoplastic liver growth. Prog Liver Dis. 1990;9:57–71. [PubMed] [Google Scholar]
- Gao C., Gressner G., Zoremba M., Gressner A. M. Transforming growth factor beta (TGF-beta) expression in isolated and cultured rat hepatocytes. J Cell Physiol. 1996 Jun;167(3):394–405. doi: 10.1002/(SICI)1097-4652(199606)167:3<394::AID-JCP3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Greenbaum L. E., Li W., Cressman D. E., Peng Y., Ciliberto G., Poli V., Taub R. CCAAT enhancer- binding protein beta is required for normal hepatocyte proliferation in mice after partial hepatectomy. J Clin Invest. 1998 Sep 1;102(5):996–1007. doi: 10.1172/JCI3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Tugores A., Veloz L., Karin M., Brenner D. A. A simplified method for the preparation of transcriptionally active liver nuclear extracts. DNA Cell Biol. 1990 Dec;9(10):777–781. doi: 10.1089/dna.1990.9.777. [DOI] [PubMed] [Google Scholar]
- Horsmans Y., Stevens M., Geubel A., Harvengt C., Rahier J. Immunoquantification of cytochrome P-450 3A on rat paraffin-embedded liver tissue. Liver. 1992 Oct;12(5):344–350. doi: 10.1111/j.1600-0676.1992.tb00584.x. [DOI] [PubMed] [Google Scholar]
- Jakowlew S. B., Mead J. E., Danielpour D., Wu J., Roberts A. B., Fausto N. Transforming growth factor-beta (TGF-beta) isoforms in rat liver regeneration: messenger RNA expression and activation of latent TGF-beta. Cell Regul. 1991 Jul;2(7):535–548. doi: 10.1091/mbc.2.7.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaumot M., Estanyol J. M., Serratosa J., Agell N., Bachs O. Activation of cdk4 and cdk2 during rat liver regeneration is associated with intranuclear rearrangements of cyclin-cdk complexes. Hepatology. 1999 Feb;29(2):385–395. doi: 10.1002/hep.510290226. [DOI] [PubMed] [Google Scholar]
- Kawada M., Yamagoe S., Murakami Y., Suzuki K., Mizuno S., Uehara Y. Induction of p27Kip1 degradation and anchorage independence by Ras through the MAP kinase signaling pathway. Oncogene. 1997 Aug 7;15(6):629–637. doi: 10.1038/sj.onc.1201228. [DOI] [PubMed] [Google Scholar]
- Knoblich J. A., Sauer K., Jones L., Richardson H., Saint R., Lehner C. F. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994 Apr 8;77(1):107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Koff A., Cross F., Fisher A., Schumacher J., Leguellec K., Philippe M., Roberts J. M. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell. 1991 Sep 20;66(6):1217–1228. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- Kren B. T., Trembley J. H., Steer C. J. Alterations in mRNA stability during rat liver regeneration. Am J Physiol. 1996 May;270(5 Pt 1):G763–G777. doi: 10.1152/ajpgi.1996.270.5.G763. [DOI] [PubMed] [Google Scholar]
- Lambotte L., Li B., Leclercq I., Sempoux C., Saliez A., Horsmans Y. The compensatory hyperplasia (liver regeneration) following ligation of a portal branch is initiated before the atrophy of the deprived lobes. J Hepatol. 2000 Jun;32(6):940–945. doi: 10.1016/s0168-8278(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Lambotte L., Saliez A., Triest S., Tagliaferri E. M., Barker A. P., Baranski A. G. Control of rate and extent of the proliferative response after partial hepatectomy. Am J Physiol. 1997 Oct;273(4 Pt 1):G905–G912. doi: 10.1152/ajpgi.1997.273.4.G905. [DOI] [PubMed] [Google Scholar]
- Leone G., DeGregori J., Sears R., Jakoi L., Nevins J. R. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997 May 22;387(6631):422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- McIntyre M., Desdouets C., Sénamaud-Beaufort C., Laurent-Winter C., Lamas E., Bréchot C. Differential expression of the cyclin-dependent kinase inhibitor P27 in primary hepatocytes in early-mid G1 and G1/S transitions. Oncogene. 1999 Aug 12;18(32):4577–4585. doi: 10.1038/sj.onc.1202815. [DOI] [PubMed] [Google Scholar]
- Mercer W. E., Avignolo C., Baserga R. Role of the p53 protein in cell proliferation as studied by microinjection of monoclonal antibodies. Mol Cell Biol. 1984 Feb;4(2):276–281. doi: 10.1128/mcb.4.2.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos G. K., DeFrances M. C. Liver regeneration. Science. 1997 Apr 4;276(5309):60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Mittnacht S., Paterson H., Olson M. F., Marshall C. J. Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr Biol. 1997 Mar 1;7(3):219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- Mosner J., Deppert W. p53 and mdm2 are expressed independently during cellular proliferation. Oncogene. 1994 Nov;9(11):3321–3328. [PubMed] [Google Scholar]
- Nakamura T., Arakaki R., Ichihara A. Interleukin-1 beta is a potent growth inhibitor of adult rat hepatocytes in primary culture. Exp Cell Res. 1988 Dec;179(2):488–497. doi: 10.1016/0014-4827(88)90286-8. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Tomita Y., Hirai R., Yamaoka K., Kaji K., Ichihara A. Inhibitory effect of transforming growth factor-beta on DNA synthesis of adult rat hepatocytes in primary culture. Biochem Biophys Res Commun. 1985 Dec 31;133(3):1042–1050. doi: 10.1016/0006-291x(85)91241-0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Theodoras A. M., Schumacher J., Roberts J. M., Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995 May;15(5):2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeper D. S., Upton T. M., Ladha M. H., Neuman E., Zalvide J., Bernards R., DeCaprio J. A., Ewen M. E. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997 Mar 13;386(6621):177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- Polyak K., Waldman T., He T. C., Kinzler K. W., Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996 Aug 1;10(15):1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- Pronk G. J., Bos J. L. The role of p21ras in receptor tyrosine kinase signalling. Biochim Biophys Acta. 1994 Dec 30;1198(2-3):131–147. doi: 10.1016/0304-419x(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Rai R. M., Loffreda S., Karp C. L., Yang S. Q., Lin H. Z., Diehl A. M. Kupffer cell depletion abolishes induction of interleukin-10 and permits sustained overexpression of tumor necrosis factor alpha messenger RNA in the regenerating rat liver. Hepatology. 1997 Apr;25(4):889–895. doi: 10.1002/hep.510250417. [DOI] [PubMed] [Google Scholar]
- Rozga J., Jeppsson B., Bengmark S. Portal branch ligation in the rat. Reevaluation of a model. Am J Pathol. 1986 Nov;125(2):300–308. [PMC free article] [PubMed] [Google Scholar]
- Russell W. E., Coffey R. J., Jr, Ouellette A. J., Moses H. L. Type beta transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5126–5130. doi: 10.1073/pnas.85.14.5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing A., Rönicke V., Bürger C., Funk M., Müller R. Alternative splicing of human cyclin E. J Cell Sci. 1994 Feb;107(Pt 2):581–588. doi: 10.1242/jcs.107.2.581. [DOI] [PubMed] [Google Scholar]
- Shaulsky G., Ben-Ze'ev A., Rotter V. Subcellular distribution of the p53 protein during the cell cycle of Balb/c 3T3 cells. Oncogene. 1990 Nov;5(11):1707–1711. [PubMed] [Google Scholar]
- Sherr C. J. Mammalian G1 cyclins. Cell. 1993 Jun 18;73(6):1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Sherr C. J., Roberts J. M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999 Jun 15;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Shi S. R., Key M. E., Kalra K. L. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 1991 Jun;39(6):741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- Simpson K. J., Lukacs N. W., Colletti L., Strieter R. M., Kunkel S. L. Cytokines and the liver. J Hepatol. 1997 Dec;27(6):1120–1132. doi: 10.1016/s0168-8278(97)80160-2. [DOI] [PubMed] [Google Scholar]
- Soussi T., Caron de Fromentel C., Breugnot C., May E. Nucleotide sequence of a cDNA encoding the rat p53 nuclear oncoprotein. Nucleic Acids Res. 1988 Dec 9;16(23):11384–11384. doi: 10.1093/nar/16.23.11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain A. J., Frazer A., Hill D. J., Milner R. D. Transforming growth factor beta inhibits DNA synthesis in hepatocytes isolated from normal and regenerating rat liver. Biochem Biophys Res Commun. 1987 May 29;145(1):436–442. doi: 10.1016/0006-291x(87)91340-4. [DOI] [PubMed] [Google Scholar]
- Stärkel P., Horsmans Y., Sempoux C., De Saeger C., Wary J., Lause P., Maiter D., Lambotte L. After portal branch ligation in rat, nuclear factor kappaB, interleukin-6, signal transducers and activators of transcription 3, c-fos, c-myc, and c-jun are similarly induced in the ligated and nonligated lobes. Hepatology. 1999 May;29(5):1463–1470. doi: 10.1002/hep.510290503. [DOI] [PubMed] [Google Scholar]
- Sugiyama A., Nagaki M., Shidoji Y., Moriwaki H., Muto Y. Regulation of cell cycle-related genes in rat hepatocytes by transforming growth factor beta1. Biochem Biophys Res Commun. 1997 Sep 18;238(2):539–543. doi: 10.1006/bbrc.1997.7338. [DOI] [PubMed] [Google Scholar]
- Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996 Mar;10(4):413–427. [PubMed] [Google Scholar]
- Thompson N. L., Mead J. E., Braun L., Goyette M., Shank P. R., Fausto N. Sequential protooncogene expression during rat liver regeneration. Cancer Res. 1986 Jun;46(6):3111–3117. [PubMed] [Google Scholar]
- Thoresen G. H., Refsnes M., Christoffersen T. Inhibition of hepatocyte DNA synthesis by transforming growth factor beta 1 and cyclic AMP: effect immediately before the G1/S border. Cancer Res. 1992 Jul 1;52(13):3598–3603. [PubMed] [Google Scholar]
- Trautwein C., Will M., Kubicka S., Rakemann T., Flemming P., Manns M. P. 2-acetaminofluorene blocks cell cycle progression after hepatectomy by p21 induction and lack of cyclin E expression. Oncogene. 1999 Nov 11;18(47):6443–6453. doi: 10.1038/sj.onc.1203045. [DOI] [PubMed] [Google Scholar]
- Tsai L. H., Lees E., Faha B., Harlow E., Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993 Jun;8(6):1593–1602. [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. Q., Lin H. Z., Yin M., Albrecht J. H., Diehl A. M. Effects of chronic ethanol consumption on cytokine regulation of liver regeneration. Am J Physiol. 1998 Oct;275(4 Pt 1):G696–G704. doi: 10.1152/ajpgi.1998.275.4.G696. [DOI] [PubMed] [Google Scholar]