Abstract
A technique has been developed to rapidly screen enzyme inhibitor candidates from complex mixtures, such as those created by combinatorial synthesis. Inhibitor libraries are screened by using immobilized enzyme technologies and electrospray ionization ion cyclotron resonance mass spectrometry. The library mixture is first sprayed into the mass spectrometer, and compounds are identified. The library is subsequently incubated with the immobilized enzyme of interest under the correct conditions (buffer, pH, temperature) by using an excess of enzyme to ensure a surplus of sites for ligand binding. The immobilized enzyme/inhibitor mixture is centrifuged, and an aliquot of supernatant is again analyzed by electrospray ionization mass spectrometry. Potential inhibitors are quickly identified by comparison of the spectra before and after incubation with the immobilized enzyme. Non-inhibitors show no change in ion intensity after incubation, whereas weak inhibitors exhibit a visible decrease in ion abundance. Once inhibitor candidates have been identified, the library is reinjected into the mass spectrometer, and tandem mass spectrometry is used to determine the structure of the inhibitor candidates as needed. This method has been successfully demonstrated by identifying inhibitors of the enzymes pepsin and glutathione S-transferase from a 19- and 17-component library, respectively. It is further shown that the immobilized enzyme can be recycled and reused for continuous screening of additional new libraries without adding additional enzyme.
During the last decade new combinatorial techniques have been developed that allow for synthesis of vast quantities of potential therapeutic compounds (1–4). Although remarkable advances in generating complex libraries of molecules have been made, the analytical process of analyzing and screening the immense numbers of compounds generated is currently lacking. To keep pace with the synthetic process, contemporary analytical techniques must be capable of screening a large number of compounds in a high-throughput manner with precision and accuracy.
Notable progress has been made in overcoming the problem of screening extensive libraries that are created by combinatorial methods using MS (5–8). At present, a variety of front-end affinity-selection techniques are used in conjunction with MS to determine potentially active compounds. Although the nomenclature of the techniques may be different, the solution phase screening methods are all generally based on the same principles. The screening process is initiated by forming a protein/ligand complex, followed by isolation of the complex by using, for example, size exclusion chromatography (9–12) or a molecular weight cut-off membrane (13, 14). Determination of bound ligands requires dissociation of the complex, followed by chromatographic-mass spectrometric detection. Some examples include pulsed ultrafiltration MS, developed by van Breemen and coworkers, as an elegant combinatorial library screening methodology (13, 15). There are also a variety of screening procedures based on frontal affinity chromatography-MS (16–20) and capillary electrophoresis-MS (21, 22).
The work described herein introduces an immobilized enzyme affinity selection procedure followed by electrospray ionization Fourier transform ion cyclotron resonance MS (ESI FT-ICR) as an inhibitor screening assay. This immobilized enzyme MS (IEMS) screening technique does not require dissociation or separation of the protein/ligand complex for identification of strong inhibitor candidates, and chromatography is not necessary because all isolations are performed in the gas-phase by using ion trap capabilities. The use of FT-ICR as the detection system in the analysis of complex libraries offers considerable advantages compared with other mass spectrometers (23, 24). The exact mass measurements, high resolution, and multiple stage mass spectrometry (MSn) capabilities facilitate the elucidation of complex mixtures, such as those generated from combinatorial libraries. Mass accuracy to within 1.5 ppm and resolution of nominal mass isobars routinely yield unambiguous information as to composition and structure of individual components from such complex mixtures.
The IEMS assay for screening mixtures of potential binding ligands and inhibitors is performed by analyzing the library before and after incubation with the immobilized enzyme by using ESI FT-ICR mass spectrometry. Potential inhibitors are quickly identified by comparison of the spectra before and after incubation with the immobilized enzyme; i.e., relative ion abundance of inhibitors decreases or disappears after incubation because of binding with the enzyme. Non-inhibitors show no change in ion intensity after incubation, whereas the ion abundance of very strong inhibitors disappears completely. Ion intensities for moderate or weak inhibitors decrease noticeably. To simplify the data handling, spectra subtraction of the original before and after incubation libraries quickly identifies all strong potential inhibitors. Once inhibitor candidates have been identified, the final step is to reinject the library into the mass spectrometer, isolate the molecular ion of the potential inhibitor candidates in the gas-phase, and use collision-induced dissociation to determine the structure. The technique can be used to rapidly identify and structurally characterize potential inhibitors from complex solution phase libraries for protein/ligand interactions without the need to isolate and dissociate enzyme/inhibitor complexes in the solution phase.
Using a molar excess of immobilized enzyme compared with the total molar concentration of the library is essential for the success of the screening assay. Excess enzyme ensures ample binding sites for the competitive inhibitors during incubation, which leads to the complete loss of ion intensity of strong binding ligands in the mass spectrum. The need for excess enzyme is offset by the ability to regenerate the active immobilized enzyme after the screening assay. Immobilized enzymes are easily recycled by denaturing the enzyme to release bound inhibitors, followed by refolding the enzyme in the proper solution environment. Overall, this recycling allows for multiple screening analyses with minimal amounts of target enzyme. It is important to acknowledge the possibility that immobilized enzymes may have different affinity characteristics from the native solution phase conformation. At this point in time, we show proof of principle that the ligands do bind to the immobilized enzyme and that activity is retained as observed by the depletion of inhibitor molecular ions in the assay.
To validate the technique, two model enzymatic systems were chosen. The enzymes pepsin and glutathione S-transferase (GST) were immobilized, and libraries of potential inhibitors were screened by using the described IEMS assay. Binding ligands for the two enzymes were rapidly and correctly identified by using this technique.
Experimental Procedures
Reagents.
Pepsin (from porcine stomach mucosa), GST (from equine liver), sodium cyanoborohydride, and all library components were purchased from Sigma. Ethanolamine was obtained from Aldrich. Aminolink coupling gel (4% cross-linked beaded agarose, 50% slurry) was purchased from Pierce.
Enzyme Immobilization.
Enzyme immobilization was performed through reductive amination between the protein primary amines and the aldehyde functional groups of the Aminolink coupling gel. For pepsin, the coupling buffer was 0.1 M sodium phosphate buffer (pH 4.5), the blocking buffer was 1.0 M ethanolamine HCl (pH 4.4), the wash buffer was 0.1 M NaPO4/1.0 M NaCl (pH 4.5), and the incubation buffer was 0.02 M sodium acetate buffer (pH 4.0). For GST, the coupling buffer was 0.1 M sodium phosphate buffer (pH 6.8), the blocking buffer was 1.0 M ethanolamine HCl (pH 7.0), the wash buffer was 1.0 M sodium chloride, and the incubation buffer was 0.02 M ammonium acetate (pH 6.8). The enzyme (4–10 mg) was dissolved in 100 μl of coupling buffer and dialyzed on a 0.025-mm nitrocellulose drop dialysis membrane (Millipore) for 1 h over coupling buffer, then diluted to 0.5 to 2 ml in coupling buffer. The dialyzed enzyme was then added to 0.5 to 2 ml of Aminolink coupling gel slurry in an empty 5-ml polypropylene affinity column (Pierce) that was equilibrated with coupling buffer, followed by 40 to 200 μl of 1 M sodium cyano borohydride solution prepared in coupling buffer. The column was capped and gently mixed overnight by using a rocking platform shaker. The column was drained and washed with 4 ml of coupling buffer. All washes were collected for determination of enzyme immobilization yield. To block excess reactive sites on the Aminolink gel, 1 ml of blocking buffer was added to the column followed by mixing for 2 h. The column was then washed successively with 15 ml of coupling buffer, followed by 20 ml of wash buffer and finally with 20 ml incubation buffer. The immobilized enzymes may be stored in a 50% glycerol solution with coupling buffer at 4°C.
Coupling Efficiency.
Enzyme immobilization yields were determined by comparison of the amount of enzyme present before and after coupling to the gel. Free enzyme concentrations were determined by their absorbance at 280 nm by using a Uvikon 933 double beam UV/VIS spectrophotometer (Kontron Instruments, Milan, Italy). The procedures of Lowry (25) and Domen (26) were followed by using the appropriate enzyme to construct standard calibration curves.
Binding Assay.
Although the final concentration of each component during the assay was approximately 4 μM, a stock solution of each component of the pepsin library was prepared at a concentration of 0.001 M in water. The inhibitor screening assay was carried out by using 0.5 ml of immobilized enzyme gel (approximately 1.5 × 10−7 mols pepsin and 2 × 10−7 mols GST, see Results). The inhibitor libraries were prepared from 2 μl of each 0.001 M component, yielding approximately 2 × 10−9 mols of each component, totaling 3.8 × 10−8 mols of pepsin library components and 3.4 × 10−8 total mols for the GST library, thus ensuring a molar excess of enzyme to ligands. The library solution was brought up to a final volume of 500 μl in incubation buffer and thoroughly mixed, yielding a final concentration of approximately 4 μM per component. A 400-μl aliquot of the library solution was incubated with the immobilized pepsin slurry in a 1.5-ml Eppendorf tube for 1 h at room temperature with gentle rocking. The incubation mixture was centrifuged for 5 min at 3000 × g, and 50 μl of supernatant was removed for ESI-MS analysis. For the pepsin assay, the 50-μl supernatant aliquot was subjected to drop dialysis for 1 h over water by using a Millipore 0.025-mm nitrocellulose membrane to remove the sodium acetate buffer. There was no detectable loss of components from the dialysis. The sample was then added to 60 μl of MeOH for electrospray ionization.
Immobilized Enzyme Recycling.
To allow continuous screening of the immobilized receptor, the enzymes were recycled. To remove the unbound library components from the immobilized enzymes, three, 1-ml washes of incubation buffer were added to the gel followed by vortexing, centrifugation, and removal of the supernatant. The immobilized enzymes were then denatured to release the bound ligands. The denaturing wash was performed by adding two 1-ml aliquots of MeOH to the immobilized enzyme gel and heating at 65°C for 10 min, followed by centrifugation and collection of the supernatant. The MeOH washes were combined and vacuum centrifuged to dryness. For ESI, the lyophilized samples were redissolved in 50 μl of 60:40 MeOH:H2O.
Mass Spectrometry.
Spectra were acquired on an Apex II FT-ICR mass spectrometer (Bruker, Billerica, MA) equipped with a 7-T actively shielded superconducting magnet. Ions were formed in a pneumatically assisted electrospray source (Analytica, Branford, CT) employing an off-axis electrospray probe at 1 μl/min. Ions were externally accumulated in a rf-only hexapole for 2 s before transfer into the ICR cell for mass analysis. Ions of interest were isolated in the gas-phase by using a series of frequency sweeps, and single frequency shots were controlled by using correlated harmonic excitation fields (27). Ions were collisionally activated by sustained off-resonance irradiation (SORI) at 500 Hz above the cyclotron frequency for 250 ms by using Ar as the collision gas. Each spectrum is an average 20 to 50 transients composed of 512,000 data points acquired by using a Bruker data station operating XMASS version 5.01.
Results
Pepsin.
To validate the new methodology, the enzyme pepsin was chosen as the first model system. Pepsin is the principal acid protease in gastric fluids and is involved in digestion processes. The inhibition of pepsin has been thoroughly studied and, therefore, is well characterized. The sequence of pig pepsin contains only two primary amines available for reductive amination; one on Lys and the other at the N-terminal amino acid, thus reducing the probability that its native affinity characteristics may be altered after immobilization.
Before the IEMS assay was performed, the pepsin immobilization yield was determined. Two separate immobilization yields were calculated: (i) the total maximum loading capacity of pepsin on the Aminolink gel and (ii) the immobilization efficiency of pepsin. For the maximum enzyme loading determination, 40 mg of pepsin was used for the immobilization with 1.0 ml of agarose gel. The unimmobilized pepsin was determined to be 19.7 mg from the UV spectroscopy assay, which equates to approximately 10 mg of enzyme per ml of agarose gel. To determine the immobilization efficiency of pepsin through reductive amination, another immobilization was performed by using 5 mg of pepsin and 1.0 ml of Aminolink gel, ensuring the maximum loading capacity of the gel was not exceeded. The UV spectroscopy assay determined the free unimmobilized pepsin to be approximately 0.4 grams, yielding an overall immobilization of 92.4%.
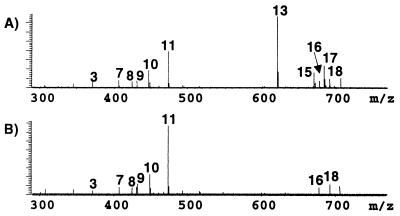
A library of compounds was assembled to test the screening capabilities of the IEMS assay (Table 1). Fig. 1A is the negative ion ESI FT-ICR mass spectrum of the 19-component library before incubation with the immobilized pepsin, and each peak is labeled according to its component number indicated in Table 1. The mass spectral analysis of the libraries must be performed in both polarities to ensure efficient ionization and detection of all components. Although spectra of each library were acquired in both the positive and negative mode, only one or the other polarity is presented for brevity. Fig. 1B is the negative ion mass spectrum of the library after a 1-h incubation period with the immobilized enzyme. Clearly, comparison of the two spectra reveals a total loss of ion intensity for known inhibitors, nos. 13, 15, and 17, which indicates binding. (The two other known inhibitors, compound nos. 6 and 11, were correctly identified as weak binding ligands, as detailed below.) In particular, compound 11 appears to increase in abundance. However, it has decreased relative to other ions in the spectra, thus signifying a weak inhibitor. N-acetyl-L-Phe-3,5-diiodo-Tyr (no. 13) has the weakest Ki at 80 μM (28), whereas the strongest binding ligand is pepstatin A (no. 17), with a Ki of 45 pm (29). These values give an estimation of the Ki's needed for determination of candidates as binding ligands by using the IEMS assay. If a component has a Ki of approximately 100 μM or lower, it may be identified as a binding ligand because of either a decrease in relative abundance or a total loss of molecular ion intensity after incubation with the immobilized enzyme.
Table 1.
Composition of library 1; known inhibitor mixture for pepsin
| Compound | Binding status* | Elemental composition | MW |
|---|---|---|---|
| 1. Diisopropyl l-tartrate | P | C10H18O6 | 234.1104 |
| 2. Tetraalanine | N | C12H22N4O5 | 302.1590 |
| 3. Bestatin | P | C16H24N2O4 | 308.1737 |
| 4. Lithocholic acid | P | C24H40O3 | 376.2978 |
| 5. Lithocholic acid methyl ester | P | C25H42O3 | 390.3134 |
| 6. Chenodeoxycholic acid | I | C24H40O4 | 392.2927 |
| 7. Cholic acid | P | C24H40O5 | 408.2876 |
| 8. Leupeptin | P | C20H38N6O4 | 426.2955 |
| 9. Arg-Gly-Asp-Ser | N | C15H27N7O8 | 433.1999 |
| 10. Glycochenodeoxycholic acid | P | C26H42NO5 | 449.3142 |
| 11. N-Acetyl-3,5-diiodo-l-Tyr | I | C11H11NO4I2 | 474.8778 |
| 12. Antipain | P | C27H44N10O6 | 604.3446 |
| 13. N-Acetyl-l-Phe-3,5-diiodo-Tyr | I | C20H20N2O5I2 | 621.9461 |
| 14. p-Aminophenylacetyl-tuftsin | N | C29H47N9O7 | 633.3599 |
| 15. Pepsinostreptin | I | C33H61N5O9 | 671.4470 |
| 16. Thymopentin | N | C30H49N9O9 | 679.3654 |
| 17. Pepstatin A | I | C43H63N5O9 | 685.4626 |
| 18. Splenopentin | N | C31H51N9O9 | 693.3811 |
| 19. Momamy peptide | N | C43H46N8O6 | 770.3841 |
I, known inhibitor; P, potential inhibitor; N, non-inhibitor.
Figure 1.
Negative mode ESI FT-ICR mass spectrum of library 1 before (A) and after (B) incubation with immobilized pepsin.
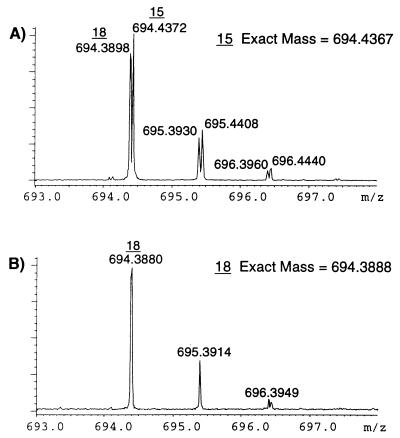
The high resolving power and utility of using FT-ICR in the IEMS assay is exemplified in the expanded positive ion spectra shown in Fig. 2 A and B. Although the sodiated ion of no. 15 is isobaric with the protonated ion of no. 18, both are clearly resolved. Comparison of the spectra before (Fig. 2A) and after (Fig. 2B) incubation with the immobilized pepsin distinctly indicates a total loss of ion intensity at m/z 694.43 (no. 15) whereas no loss of ion intensity at m/z 694.38 (no. 18) is observed. Compound 15, pepsinostreptin, is a known strong inhibitor of pepsin, whereas no. 18, splenopentin, is a non-inhibitor. Mass measurement accuracy was also such that compound 15 was measured to within 0.7 ppm of the theoretical mass whereas no. 18 was determined to within 1.4 ppm by using external calibration. Both the high resolution and accurate mass measurement capabilities increase the probability of correctly analyzing complex mixtures such as those generated from combinatorial libraries.
Figure 2.
Positive ion mode ESI FT-ICR spectrum of region around m/z 694 before (A) and after (B) incubation with immobilized pepsin.
The IEMS assay may also be used to obtain structural information by using MSn techniques. Structural characterization is performed by reinfusing the intact library into the mass spectrometer and isolating the ions of interest in the gas phase by using correlated harmonic excitation frequencies (CHEF) (27), followed by collision-induced dissociation. Although protonated splenopentin and sodiated pepsinostreptin differ by only 0.05 Da, the inhibitor was successfully isolated, demonstrating that chromatography is not necessary for the isolation and structural characterization of binding ligands from complex mixtures. The sustained off-resonance irradiation collision-induced dissociation spectrum of the [M + Na]+ ion for no. 18, along with its structure and generated fragments, was easily obtained (see supplementary Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org). Sufficient fragment ions were produced from the sodiated ion for complete characterization, demonstrating the ability of the technique to provide structural information of binding candidates.
Given the probability that only small amounts of enzyme might be available for screening, a method was devised that allows the enzyme to be reused several times. In this way, the original immobilized enzyme can be repeatedly used in screening several different libraries without consuming additional enzyme. To recycle the immobilized enzyme, it is first washed with the incubation buffer, followed by exchange of solvent to a denaturing environment (see Experimental Procedures for details). This procedure releases the binding ligands, which can be collected and analyzed. The proper enzyme environment is then regenerated by suspending it in the appropriate buffer such that the fully active immobilized enzyme can be repetitively used for screening additional libraries. To demonstrate the reliability of the recycled immobilized enzyme, the IEMS assay was repeated with the pepsin library by using the recycled pepsin. Comparison of data after recyclization reveals nearly identical spectra, indicating that the enzyme has been successfully recycled with little or no loss in affinity characteristics or activity. This method of recycling the immobilized enzyme has been successfully repeated at least five times with pepsin, thus suggesting that at least five different libraries can be screened against the original immobilized enzyme.
In addition to recycling the enzyme, an added advantage of denaturing the enzyme is the fact that both strong and weak binding components can be released and analyzed by MS. Although the recycling experiment is not necessary for the identification of strong binding ligands, it presents a simple method of distinguishing between potential tight and weak binding compounds and provides a second check on identification of strong binding ligands. Weak binding candidates are considered those that bind to the enzyme during incubation but are still present in the mass spectrum after incubation because of larger dissociation constant values. Table 2 lists all of the weak binding compounds identified by the release experiment. Compounds nos. 7, 10, and 11 were released from the enzyme along with the strong binding compounds (nos. 13, 15, and 17). Additional weakly bound compounds (nos. 1, 3, 6, and 8) were identified from the positive ion spectrum. The components bestatin (no. 3) and leupeptin (no. 8) are general protease inhibitors and were expected to bind weakly to pepsin. Cholic acid (no. 7) and glycochenodeoxycholic acid (no. 10) are derivatives of the weak inhibitor chenodeoxy acid (no. 6) and were identified as possible weak binding ligands, as was diisopropyl-L-tartrate (no. 1). N-acetyl-3,5-diiodo-L-Tyr (no. 11), a known weak inhibitor, has a Ki of 880 μM (30). Thus, both ligands with Ki's ranging from 45 pM (pepstatin A) to approximately 900 μM (N-acetyl-3,5-diiodo-L-Tyr) were successfully identified by using the release experiment, although the procedure was not a requisite to identify the strong inhibitors, as shown above.
Table 2.
Weak pepsin binding ligands identified from library 1 using recycle experiment
| Library component number | Compound |
|---|---|
| 1 | Diisopropyl l-tartrate |
| 3 | Bestatin |
| 6 | Chenodeoxycholic acid |
| 7 | Cholic acid |
| 8 | Leupeptin |
| 10 | Glycochenodeoxycholic acid |
| 11 | N-Acetyl-3,5-diiodo-l-Tyr |
Glutathione S-Transferase.
The IEMS assay was further evaluated by examining a second, more structurally complex model enzymatic system, GST. GST is a detoxifying enzyme found in various organisms and catalyzes the nucleophilic attack of glutathione to a variety of electrophilic substrates. GST is considered an important therapeutic target based, in part, on its implication in the biological resistance of certain anti-cancer drugs. The sequence of various GSTs contain about 10% lysine, furnishing abundant sites for immobilization through reductive amination. It also tests the reliability of the immobilization procedure because of the large number of free amines. An initial concern was that severe distortion of the enzyme might result from the immobilization, thus rendering the enzyme inactive, and that the affinity characteristics of the enzyme might be altered.
Initially, 7.7 mg of GST was used for the immobilization onto 1.0 ml of Aminolink slurry or 0.5 ml of gel to determine the immobilization yield. Analysis of free unimmobilized GST by UV spectroscopy at 280 nm was determined to be 0.93 mg, resulting in an 87.6% immobilization yield.
As with pepsin, a library of inhibitors was constructed for GST based on the knowledge of known inhibitors, possible inhibitors containing various structural modifications of known inhibitors and known nonbinding ligands (Table 3). The final library consisted of 17 components, ranging in molecular weight from 222 to 805 Da.
Table 3.
Composition of library 2; known inhibitor mixture for GST
| Compound | Binding status* | Elemental composition | MW | Before incubation† | After incubation† |
|---|---|---|---|---|---|
| 1. Flavone | I | C15H10O2 | 222.0681 | X | O |
| 2. S-Methylglutathione | P | C11H19N3O6S | 321.0994 | X | X |
| 3. S-Ethylglutathione | P | C12H21N3O6S | 335.1151 | X | X |
| 4. S-Propylglutathione | P | C13H23N3O6S | 349.1307 | X | X |
| 5. Glutathione sulfonic acid | P | C10H17N3O9S | 355.0685 | X | O |
| 6. S-Butylglutathione | P | C14H25N3O6S | 363.1464 | X | X |
| 7. S-(Lactoyl)-glutathione | P | C13H21N3O8S | 379.1049 | X | X |
| 8. S-Hexylglutathione | I | C16H29N3O6S | 391.1777 | X | (X) |
| 9. S-(p-Nitrobenzyl)-glutathione | P | C17H22N4O8S | 442.1158 | X | (X) |
| 10. S-(p-Chlorophenylacetyl)-glutathione | P | C18H22N3O7SCl | 449.3142 | X | O |
| 11. β-Casomorphin fragment | N | C30H37N5O7 | 579.2693 | X | X |
| 12. Glutathione (oxidized form) | P | C20H32N6O12S2 | 612.1519 | X | X |
| 13. Phe-Leu-Glu-Glu-Ile | N | C31H47N5O10 | 649.3323 | X | X |
| 14. Momany peptide | N | C43H46N8O6 | 770.3841 | X | (X) |
| 15. l-Thyroxine | I | C15H11NO4I4 | 776.6867 | X | O |
| 16. l-Thyroxine methyl ester | P | C16H13NO4I4 | 790.7023 | X | O |
| 17. l-Thyroxine ethyl ester | P | C17H15NO4I4 | 804.7179 | X | O |
I, known inhibitor; P, potential inhibitor; N, non-inhibitor.
X, ion present in mass spectrum; O, total loss of ion intensity after incubation with immobilized enzyme; (X), >40% decrease in ion intensity after incubation with immobilized enzyme.
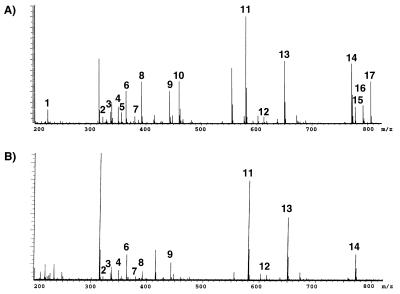
The positive ion ESI FT-ICR mass spectrum of the 17 component GST inhibitor library before incubation with immobilized GST is shown in Fig. 3A. All 17 components are clearly resolved and identified. Fig. 3B is the mass spectrum of the library after incubation for 1 h with immobilized GST. Comparison of Fig. 3 A and B indicates that components 1, 5, 10, 15, 16, and 17 have lost all ion intensity and are therefore identified as possible strong binding candidates. Ions at m/z 316 and 557 are due to ammonium acetate clusters that occur during the electrospray process. Flavone (no. 1) and L-thyroxine (no. 15) are reported strong inhibitors for GST. The published Ki for L-thyroxine is 6.6 μM (31), again supporting the claim that compounds whose dissociation constants are in the range of low pM to high mM may be assayed as binding ligands. The screening analysis also identified glutathione sulfonic acid (no. 5) and S-(p-chlorophenylacetyl)-glutathione (no. 10) as strong binding candidates. These two components are derivatives of GST's natural substrate, glutathione; therefore, their affinity for GST is not surprising. Components 16 and 17 are L-thyroxine methyl ester and L-thyroxine ethyl ester, respectively, and have not been previously tested as inhibitor candidates for GST. All strong binding ligands identified by the IEMS screening assay are shown in Table 4. The results also indicate that, even though GST is comprised of approximately 10% lysine, its binding characteristics are still retained after the immobilization. The ability of the assay to quickly identify unknown binding ligands from complex libraries is clearly an advantage in characterizing the binding properties of enzymes. For example, when the carboxylic acid of L-thyroxine was derivatized to a methyl or ethyl ester, the ligand still possessed strong binding characteristics. The ability to quickly determine, by mass analysis, binding characteristics based on differences in functional groups is an added advantage to those researchers in the area of combinatorial synthesis of enzyme inhibitors.
Figure 3.
Positive mode ESI FT-ICR mass spectrum of library 2 before (A) and after (B) incubation with immobilized GST.
Table 4.
Strong GST binding ligands identified from library 2 using IEMS assay
| Library component number | Compound |
|---|---|
| 1 | Flavone |
| 5 | Glutathione sulfonic acid |
| 10 | S-(p-chlorophenylacetyl)-glutathione |
| 15 | l-thyroxine |
| 16 | l-thyroxine methyl ester |
| 17 | l-thyroxine ethyl ester |
Table 3 also displays the IEMS assay results of the inhibitor library before and after incubation with immobilized GST in the positive ion mode. Components present in the mass spectrum are indicated by an X, components with loss of total ion intensity are represented by a 0, and components with a decrease in ion intensity (>40%) are designated by (X). The value of 40% was chosen because of possible fluctuations in ion intensities from scan to scan because of the electrospray ionization process. Components 8 (S-hexylglutathione), 9 (S-(p-nitrobenzyl)-glutathione), and 14 (momany peptide) lost greater than 40% ion intensity and may be classified as possible weak binding ligands toward GST.
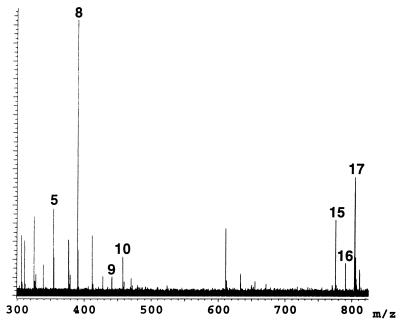
The binding components of the library were released from the immobilized GST according to the recycle experimental procedures. Fig. 4 is the negative mode ESI FT-ICR mass spectrum of the released components. Five candidates that were initially identified as strong binding components are present, reconfirming their binding affinity to GST, along with the weak binding ligands 8 and 9. The weak binding candidates were initially identified from a decrease in ion intensity after incubation with the immobilized GST and are now confirmed from the recycling experiment. S-Hexylglutathione is a known weak inhibitor for GST and was properly identified by the assay whereas S-(p-nitrobenzyl)-glutathione was identified as a new weak binding candidate.
Figure 4.
Negative ion ESI FT-ICR mass spectrum of library 2 components released from the immobilized GST employing the recycle procedure.
In addition to the two model systems discussed herein, studies have begun in the search for inhibitors for the poorly characterized enzyme class, the estrogen sulfotransferases. At this time, combinatorial libraries of possible inhibitors are being screened by using the IEMS assay and compared with a more traditional TLC assay. Thus far, identical compounds have been found to bind to the enzyme by using both assays when performed in double blind studies.
Nonspecific Binding.
The Aminolink gel was tested to determine whether nonspecific binding was occurring between the ligands and the agarose. The gel was first deactivated by blocking the reactive aldehyde groups with ethanolamine. Both the pepsin and GST libraries were individually incubated with the deactivated agarose for 1 h and centrifuged, and the supernatant was electosprayed after drop dialysis. As expected, all hydrophilic ligands were present in the mass spectrum, which strongly suggests that there were no detrimental interactions with the agarose. It has also been previously reported that nonspecific binding to Aminolink coupling gel is minimal (26). Unfortunately some of the hydrophobic components were detected in the mass spectrum after executing the recycling procedure with the deactivated agarose. Minute amounts of the components momamy peptide, lithocholic acid, and lithocholic acid methyl ester were detected. These components, which are only moderately soluble in water, are soluble in MeOH and were most likely redissolved during the recycling of the deactivated agarose. This effect presents a limitation to the procedure, in that hydrophobic components may result in false weak inhibitor hits. Hydrophobic compounds have also been encountered as problematic in other affinity-based mass spectrometric screening procedures (13). To alleviate the problem, component libraries should be soluble in the proper buffer conditions required for specific enzyme activity. Our preliminary attempt was to use agarose because it is known to reduce nonspecific binding; however, other resins or microbeads could easily be implemented instead.
Conclusions
The method of combining immobilized enzymes for the affinity-based screening of possible inhibitor libraries in conjunction with MS has been shown capable of identifying strong binding targets from complex mixtures. Use of FT-ICR mass spectrometry ensures accurate detection of inhibitor candidates because of the exact mass measurement, high resolution, and MSn capabilities. Total analysis time including a 1-h incubation is approximately 1.25 to 1.5 h. Chromatographic separation of inhibitors was not necessary given the gas-phase isolation capabilities of the instrument. The IEMS assay for screening mixtures of potential binding ligands and inhibitors is performed by analyzing the library before and after incubation with the immobilized enzyme. Potential inhibitors are quickly identified by comparison of the spectra before and after incubation with the immobilized enzyme; i.e., relative abundance of inhibitors decreases or disappears after incubation because of binding with the enzyme. Two model enzymatic systems, pepsin and GST, were used to validate the technique by screening assembled libraries of potential inhibitors. For pepsin, all strong inhibitors contained within the library were identified, and the ability to structurally characterize the ligands by using tandem mass spectrometry (MS/MS) was demonstrated. All known strong inhibitors, from a library composed of known, potential and non-inhibitors for GST, were also successfully identified. For both enzymatic models, weak binding ligands were determined from either a decrease in ion intensity after incubation or from detection of the compound after release during the enzyme recycling procedure. Immobilized enzymes were efficiently recycled for continuous inhibitor screening such that at least five different libraries could be screened by using one immobilization of pepsin. Overall, the screening technique was able to accurately detect and differentiate strong and weak binding ligands from a pool of potential target compounds, such as those created from combinatorial libraries. Currently, the IEMS assay is only proven to detect any and all ligand binding, not site-specific inhibitors. However, Ki and IC50 have been determined for compounds correctly identified as inhibitors for estrogen sulfotransferase by using this mass spectrometric methodology and verified with more traditional methods (32).
Supplementary Material
Acknowledgments
We thank Dr. Carolyn Bertozzi for use of the UV spectrophotometer. We acknowledge University of California at Berkeley unrestricted funds for financial support.
Abbreviations
- ESI FT-ICR
electrospray ionization Fourier transform ion cyclotron resonance
- IEMS
immobilized enzyme MS
- GST
glutathione S-transferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220403997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220403997
References
- 1.Thompson L A, Ellman J A. Chem Rev. 1996;96:555–600. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- 2.Janda K D. Proc Natl Acad Sci USA. 1994;91:10779–10785. doi: 10.1073/pnas.91.23.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon E M, Gallop M A, Patel D V. Acc Chem Res. 1996;29:144–154. [Google Scholar]
- 4.Van Hijfte L, Marciniak G, Froloff N. J Chromatogr B. 1999;725:3–15. doi: 10.1016/s0378-4347(99)00007-9. [DOI] [PubMed] [Google Scholar]
- 5.Submuth R D, Jung G. J Chromatogr B. 1999;725:49–65. doi: 10.1016/s0378-4347(98)00513-1. [DOI] [PubMed] [Google Scholar]
- 6.Swali V, Langley G J, Bradley M. Curr Opin Chem Biol. 1999;3:337–341. doi: 10.1016/s1367-5931(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 7.Siuzdak G, Lewis J K. Biotech Bioeng. 1998;61:127–134. [PubMed] [Google Scholar]
- 8.Woodbury C P, Venton D L. J Chromatogr B. 1999;725:113–137. doi: 10.1016/s0378-4347(99)00006-7. [DOI] [PubMed] [Google Scholar]
- 9.Dunayevskiy Y M, Lai J-J, Quinn C, Talley F, Vouros P. Rapid Commun Mass Spectrom. 1997;11:1178–1184. [Google Scholar]
- 10.Siegel M M, Tabei K, Bebernitz G A, Baum E Z. Rapid Commun Mass Spectrom. 1998;33:264–273. doi: 10.1002/(SICI)1096-9888(199803)33:3<264::AID-JMS629>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Davis R G, Anderegg R J, Blanchard S G. Tetrahedron. 1999;55:11653–11667. [Google Scholar]
- 12.Blom K F, Larsen B S, McEwen C N. J Comb Chem. 1999;1:82–90. doi: 10.1021/cc980010k. [DOI] [PubMed] [Google Scholar]
- 13.van Breemen R B, Huang C-R, Nikolic D, Woodbury C P, Zhao Y-Z, Venton D L. Anal Chem. 1997;69:2159–2164. doi: 10.1021/ac970132j. [DOI] [PubMed] [Google Scholar]
- 14.Wieboldt R, Zweigenbaum J, Henion J. Anal Chem. 1997;69:1683–1691. doi: 10.1021/ac9610265. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y-Z, van Breemen R B, Nikolic D, Huang C-R, Woodbury C P, Schilling A, Venton D L. J Med Chem. 1997;40:4006–4012. doi: 10.1021/jm960729b. [DOI] [PubMed] [Google Scholar]
- 16.Schriemer D C, Bundle D R, Li L, Hindsgaul O. Angew Chem Int Ed Engl. 1998;37:3383–3387. doi: 10.1002/(SICI)1521-3773(19981231)37:24<3383::AID-ANIE3383>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 17.Kaur S, McGuire L, Tang D, Dollinger G, Huebner V. J Protein Chem. 1997;16:505–511. doi: 10.1023/a:1026369729393. [DOI] [PubMed] [Google Scholar]
- 18.Kelly M A, Liang H, Sytwu I-I, Vlattas I, Lyons N L, Bowen B R, Wennogle L P. Biochemistry. 1996;35:11747–11755. doi: 10.1021/bi960571x. [DOI] [PubMed] [Google Scholar]
- 19.Cai J, Henion J. Anal Chem. 1996;68:72–78. doi: 10.1021/ac950763i. [DOI] [PubMed] [Google Scholar]
- 20.Nedved M L, Habibi-Goudarzi S, Ganem B, Henion J D. Anal Chem. 1996;68:4228–4236. doi: 10.1021/ac9603035. [DOI] [PubMed] [Google Scholar]
- 21.Chu Y-H, Dunayevskiy Y M, Kirby D P, Vouros P, Karger B L. J Am Chem Soc. 1996;118:7827–7835. [Google Scholar]
- 22.Lyubarskaya Y V, Carr S A, Dunnington D, Prichett W P, Fisher S M, Appelbaum E R, Jones C S, Karger B L. Anal Chem. 1998;70:4761–4770. doi: 10.1021/ac980330q. [DOI] [PubMed] [Google Scholar]
- 23.Nawrocki J P, Wigger M, Watson C H, Hayes T W, Senko M W, Benner S A, Eyler J R. Rapid Commun Mass Spectrom. 1996;10:1860–1864. doi: 10.1002/(SICI)1097-0231(199611)10:14<1860::AID-RCM770>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Wigger M, Nawrocki J P, Watson C H, Eyler J R, Benner S A. Rapid Commun Mass Spectrom. 1997;11:1749–1752. doi: 10.1002/(SICI)1097-0231(19971030)11:16<1749::AID-RCM91>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 25.Lowry O H, Rosebrough N J, Farr L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.Domen P L, Nevens J R, Mallia K, Hermanson G T, Klenk D C. J Chromatogr. 1990;510:293–302. doi: 10.1016/s0021-9673(01)93763-x. [DOI] [PubMed] [Google Scholar]
- 27.deKoning L J, Nibbering N M M, van Orden S L, Laukien F H. Int J Mass Spectrom Ion Processes. 1997;165:209–219. [Google Scholar]
- 28.Jackson W T, Schlamowitz M, Shaw A. Biochemistry. 1965;4:1537–1539. doi: 10.1021/bi00884a012. [DOI] [PubMed] [Google Scholar]
- 29.Rich D, Bernatowiczm M, Agarwal N, Kawai M, Salituro F, Schmidt P. Biochemistry. 1985;24:3165–3173. doi: 10.1021/bi00334a014. [DOI] [PubMed] [Google Scholar]
- 30.Silver M S, Denburg J L, Steffens J J. J Am Chem Soc. 1965;87:886–889. doi: 10.1021/ja01082a032. [DOI] [PubMed] [Google Scholar]
- 31.Young P R, Briedis A V. Biochim Biophys Acta. 1990;1038:114–118. doi: 10.1016/0167-4838(90)90018-b. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong J I, Portley A R, Chang Y T, Nierengarten D M, Cook B N, Bowman K G, Gray N S, Schultz P G, Bertozzi C R. Angew Chem Int Ed Engl. 2000;39:1303–1306. doi: 10.1002/(sici)1521-3773(20000403)39:7<1303::aid-anie1303>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.