Abstract
Thrombin orchestrates cellular events after injury to the vascular system and extravasation of blood into surrounding tissues. The pathophysiological response to thrombin is mediated by protease-activated receptor-1 (PAR-1), a seven-transmembrane G protein-coupled receptor expressed in the nervous system that is identical to the thrombin receptor in platelets, fibroblasts, and endothelial cells. Once activated by thrombin, PAR-1 induces rapid and dramatic changes in cell morphology, notably the retraction of growth cones, axons, and dendrites in neurons and processes in astrocytes. The signal is conveyed by a series of localized ATP-dependent reactions directed to the actin cytoskeleton. How cells meet the dynamic and localized energy demands during signal transmission is unknown. Using the yeast two-hybrid system, we identified an interaction between PAR-1 cytoplasmic tail and the brain isoform of creatine kinase, a key ATP-generating enzyme that regulates ATP within subcellular compartments. The interaction was confirmed in vitro and in vivo. Reducing creatine kinase levels or its ATP-generating potential inhibited PAR-1-mediated cellular shape changes as well as a PAR-1 signaling pathway involving the activation of RhoA, a small G protein that relays signals to the cytoskeleton. Thrombin-stimulated intracellular calcium release was not affected. Our results suggest that creatine kinase is bound to PAR-1 where it may be poised to provide bursts of site-specific high-energy phosphate necessary for efficient receptor signal transduction during cytoskeletal reorganization.
Protease activated receptor-1 (PAR-1) mediates the cellular responses to thrombin during blood coagulation, cell proliferation, vascular permeability changes, tumor metastasis, and nervous system injury (1–3). PAR-1 is a seven-transmembrane G protein-coupled receptor with a novel activation mechanism. Proteolysis at a thrombin cleavage site in the extracellular amino terminus exposes a new amino terminus containing the peptide ligand SFLLRN, which binds intramolecularly to initiate intracellular signals (4). Although originally detected in platelets, endothelial cells, and fibroblasts, PAR-1 is also expressed in the nervous system in a developmentally regulated manner and by specific subpopulations of neurons and astrocytes that are especially vulnerable to neurodegeneration and ischemic injury (1, 5, 6).
In most cells expressing PAR-1, activation of the receptor transmits signals to the actin cytoskeleton that profoundly alter cell shape. Platelets, for example, convert from a spherical to disk shape and extend filopodia (7), endothelial cells contract (8), neurons retract axons, and astrocytes resorb processes and flatten their cell bodies (9–11). These signals also regulate changes in actin-related cell motility observed in neurons (10), fibroblasts (12), and tumor cells (3). The morphological response is mediated by a key signaling pathway that uses serine/threonine kinases, Gα12/13, RhoA, and myosin light chain kinase; actomyosin contraction generates tension through the formation of stress fibers and focal adhesions (13–17). There are several points along this signal transduction pathway at which high-energy phosphate in the form of ATP is required. Only phosphorylated myosin is capable of interacting with actin filaments, and actin polymerization requires binding to ATP (18). Depletion of total cellular ATP or the application of drugs that block the ATP binding sites of serine–threonine kinases, for example, inhibits PAR-1-mediated shape changes (10, 19). Activation of Gα12/13 requires phosphorylation and a ready supply of GTP (20). Thus, the actomyosin contraction underlying cellular shape changes is an active, energy-consuming process where localized ATP homeostasis is critical (18, 21).
Creatine kinase enzymes regulate ATP homeostasis in subcellular compartments by the transfer phosphates between creatine and adenine nucleotides. The mitochondrial isoform generates creatine phosphate, which is shuttled to cytosolic isoforms strategically localized to specific subcellular regions where they provide ATP at sites of high and fluctuating energy demand (22). In sea urchin sperm, for example, mitochondrial creatine kinase is restricted to sperm heads whereas cytosolic creatine kinase is expressed in the sperm tail, and specifically inhibiting ATP generation by the cytosolic creatine kinase results in defective tail movements and sperm motility (23). The cytosolic muscle isoform localizes along the M-line to the myosin heads where it provides ATP during actomyosin contraction (24). The cytosolic brain isoform (CKB) and its insect homolog arginine kinase localize to the membranes of neurons and astrocytes and are concentrated along axons and moving lamellipodial edges of migrating glia, although the precise function of CKB is not known (25, 26).
We hypothesized that molecules necessary for the early stages of thrombin signaling might bind to the intracellular face of PAR-1. To examine this, we performed a yeast two-hybrid screen and discovered an interaction between the PAR-1 cytoplasmic tail (C-tail) and CKB. Because the muscle isoform of creatine kinase delivers a burst of compartmentalized ATP for contracting skeletal muscle and is linked to actomyosin function in various cells (22, 26, 27), we hypothesized that CKB may perform a similar function during PAR-1-directed cytoskeletal reorganization. Using three different methods to ablate CKB activity, we found that CKB was necessary for PAR-1 signal transduction to the cytoskeleton.
Methods
Materials.
The protein–protein interaction screen was performed by using the Matchmaker GAL4 two-hybrid system and a rat brain cDNA library (CLONTECH). Rabbit anti-PAR-1 antibody was a gift from M. Runge (University of Texas Medical Branch, Galveston), anti-CKB antibodies were obtained from Chemicon, and anti-RhoA was from Santa Cruz Biotechnology. CKB activity was measured with CK 520 Sigma. ATP levels were measured with the bioluminescent somatic cell assay (Sigma FL-ASC). Thrombin and purified CKB were purchased from Calbiochem, and cyclocreatine was purchased from Sigma. Site-directed mutagenesis was performed with Quick Change (Stratagene). G. Molloy (Delaware University) graciously provided the CKB plasmid, and T. Ma (Baylor College, Houston) provided the dominant negative CKB mutant. Statistical analysis was carried out by using two-way ANOVA.
Yeast Two-Hybrid Interaction.
The PCR product of rat PAR-1 C-tail (residues 382–431) and oligonucleotides for human PAR-1 cytoplasmic loops CL-1 (residues 119–135), CL-2 (residues 199–218), and CL-3 (residues 289–311) were cloned into the GAL4 DNA-binding (DB) vector pAS2–1. These bait plasmids were used in a yeast two-hybrid screen to probe a rat brain cDNA library fused to a GAL4 activation domain (AD) in pGAD10 by cotransformation into Y190 yeast. Positive clones were selected on Leu−/Trp−/His− plates and by detection of β-galactosidase activity using a filter lift method (CLONTECH). Interactions were confirmed by retransforming positive clones either with bait vector or the original pAS2–1 vector (backbone) into yeast.
Glutathione S-Transferase Pull-Down Assays.
The rat PAR-1 C-tail was subcloned into a GST vector, and the mutant fusions were constructed by introducing stop codons D400Z, Q409Z, and H421Z. Fusion proteins were purified by using glutathione–Sepharose beads (Amersham Pharmacia). CKB truncation mutants were constructed by introducing stop codons into pGEM-1/rat CKB at amino acids Q185Z, K298Z, and K358Z. CKB mutant 186–297 was generated by PCR and cloned into pGEM-1. CKB constructs were expressed and labeled with [35S]methionine/cysteine (Amersham Pharmacia) by using a rabbit reticulocyte lysate expression system (TNT T7, Invitrogen). Binding studies were performed overnight at 4°C with 25 μl of washed glutathione beads and 30–50 μg of either GST or GST–PAR-1 C-tail fusion protein in 200 μl of 25 mM Hepes, pH 7.5, 100 mM NaCl, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol, 1% Nonidet P-40, 0.2% BSA. Binding products were separated by SDS/PAGE and analyzed by autoradiography or immunoblots with anti-CKB antibodies and enhanced chemiluminescence (Amersham Pharmacia).
Coimmunoprecipitation.
Primary cortical astrocyte cultures were prepared from 1- to 2-day-old Sprague–Dawley rats as described (2). Astrocytes were solubilized and incubated with control antibody (rabbit IgG) or anti-PAR-1 antibody overnight at 4°C in the presence of protein A–Sepharose. Immunoprecipitation products were either subjected to CKB activity assays or separated by SDS/PAGE and analyzed on immunoblots with anti-CKB antibodies.
Astrocyte Morphology Studies.
Oligonucleotides based on a CKB antisense sequence (GUGUCUGCAACAGCUUAGC-CAUCAG), a control reverse sequence (GACUACCGAUUCGACAACGUCUGUG), and an identical antisense sequence rendered insensitive to RNaseH were purchased from Sequitur (Natick, MA) and transformed into primary astrocyte cultures with Lipofectin (GIBCO/BRL) for 1 h. After 18 h, cells were differentiated in 1.5 mM dibutyryl cAMP for 2 h. A total of 10 pM thrombin was added for 2 h for a maximal response, and the morphology of approximately 200 cells was examined under phase-contrast micrography and scored as either stellate or flat. Antisense knockdown of CKB was confirmed by immunoblot analysis. Morphology assays were also performed after a 4-h treatment with 50 mM cyclocreatine.
Dominant Negative CKB Studies.
The dominant negative CKB mutant C283S (28) was cloned into the pIRES-EGFP vector (CLONTECH) and transfected for 6 h with Novafector (VennNova, LCC) into Ad12HER10 retinoblasts seeded into Lab-Tek II slides (Nunc). After 18 h, cells were differentiated with 2 mM dibutyryl cAMP in serum-free medium for 6 h. After addition of thrombin (160 nM) for 2 h, transfected cells (20–30%) were identified by fluorescence micrography, and the cell morphology of approximately 100 transfected cells was examined with phase-contrast illumination and scored blind to treatment as either refractile or flat.
RhoA Activation and Translocation Assay.
Active RhoA levels were measured in primary rat astrocytes as described (29). Astrocytes received a 4-h 50 mM cyclocreatine treatment before the thrombin (10 pM) addition for 10 min. Cell lysates were incubated with GST–Rhotekin binding domain (20–30 μg) for 45 min, washed, and immunoprobed with anti-RhoA antibody. For membrane RhoA, crude membrane fractions were prepared as described (2). Fifteen micrograms of membrane protein fractions was separated by SDS/PAGE and immunoblotted with anti-RhoA antibody.
Intracellular Calcium Assay.
Astrocytes were treated with or without 50 mM cyclocreatine for 4 h. Cell medium was decanted and replaced with Fluo-3 AM (Molecular Probes) for 1 h. Some wells received 1 μM U-73122 (Biomol, Plymouth Meeting, PA) for 10 min before 10 pM thrombin and 10 mM EDTA injection. The relative intracellular calcium levels were monitored at 488 nM for 210 sec in 4–8 identically treated wells by using a Fluorometric Imaging Plate Reader (Molecular Devices).
Results
PAR-1 Interacts with CKB in Vitro and in Vivo.
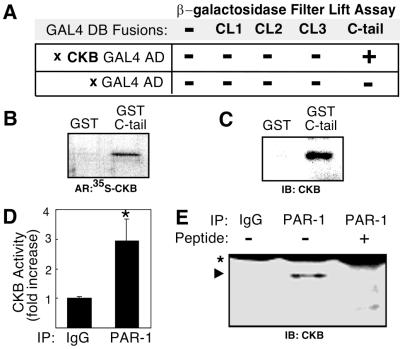
Membrane receptors depend on specific contacts selectively made with effector molecules to create intracellular signals (30). To identify molecular components required for PAR-1 signal transduction, the 51-amino acid segment corresponding to the rat PAR-1 C-tail was used to probe approximately 3.5 × 106 cDNAs from a rat brain cDNA library for binding partners in a yeast two-hybrid screen. Eight colonies representing putative interactions were detected between the PAR-1 C-tail and cDNAs in a β-galactosidase filter lift assay. Nucleotide sequencing and comparison to sequences in the GenBank database revealed that the cDNA from a single colony encoded amino acids 185–381 of CKB (22). The CKB interaction was specific to the C-tail and was not detected with CL-1, -2, -3 or with the vectors alone (Fig. 1A). This interaction was confirmed in a GST pull-down assay in which full-length 35S-labeled CKB was incubated with either GST or a GST–PAR-1 C-tail fusion protein. Analysis of binding products by SDS/PAGE and autoradiography revealed that 35S-labeled CKB bound only the GST-C-tail fusion and not GST alone (Fig. 1B). In a separate binding experiment using human brain-purified CKB, a specific interaction with GST-C-tail was also observed (Fig. 1C). The yeast two-hybrid and in vitro binding studies suggested PAR-1 may directly interact with CKB through its C-tail.
Figure 1.
CKB interaction with PAR-1. (A) Yeast two-hybrid β-galactosidase filter lift assay summary. PAR-1 cytoplasmic loops (CL1, CL2, CL3) and C-tail fused with a GAL4 DNA binding domain (DB) were coexpressed with partial cDNA clones encoding either CKB fused to a GAL4 activation domain (AD) or the AD vector alone. A positive interaction was only detected between the PAR-1 C-tail and CKB. (B) Pull-down assay shows specific interaction between 35S-labeled CKB and GST–C-tail fusion protein but not GST alone. Autoradiograph (AR). (C) Pull-down assay using purified CKB. Immunoblot (IB) with anti-CKB antibody. (D) CKB activity detection after immunoprecipitation (IP) with control IgG (lane 1) or with anti-PAR-1 antibodies (lane 2) from primary astrocyte cultures. (E) CKB protein coimmunoprecipitates with anti-PAR-1 antibodies (lane 2, arrowhead) but not control IgG (lane 1), or anti-PAR-1 antibodies preadsorbed with antigenic peptide SFLLRN (lane 3). Ig heavy chain (*).
To determine whether PAR-1 was bound to CKB under physiological conditions, we tested whether PAR-1 antibodies would co-immunoprecipitate CKB from primary cultures of rat astrocytes. Antibody complexes were affinity purified and analyzed for the presence of CKB using a highly specific and sensitive CKB colorimetric enzyme assay. We detected more than a 3-fold higher level of CKB activity in PAR-1 immunoprecipitates as compared with control IgG precipitates (Fig. 1D). To further confirm the interaction, PAR-1 immunoprecipitates were separated by SDS/PAGE and analyzed for the presence of CKB. Immunoblots against CKB demonstrated that anti-PAR-1 antibodies specifically coprecipitated CKB because CKB did not coprecipitate with control serum or with anti-PAR-1 antibodies preadsorbed with its antigenic SFLLRN peptide (Fig. 1E). The CKB signal comigrated with the CKB band detected in rat brain extracts (data not shown).
Mutational Analysis Identifies PAR-1 and CKB Interaction Domains.
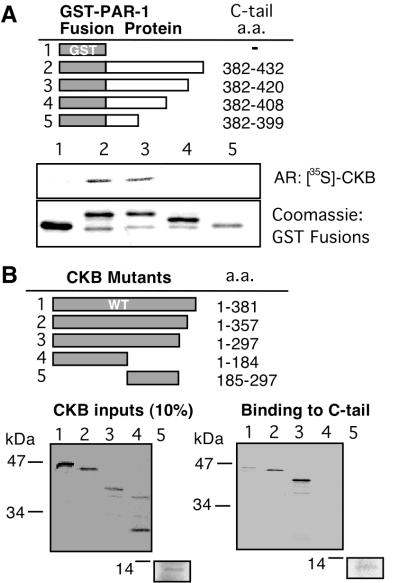
To identify segments of the C-tail required for the CKB interaction, GST fusions to PAR-1 C-tail truncation mutants were generated. These mutants were analyzed for their ability to bind metabolically labeled CKB. 35S-labeled CKB bound C-tail mutant 382–420 in which the last 12 amino acids were removed. Binding was abolished, however, with further deletions in C-tail mutants 382–408 and 382–399. This suggests that the intervening C-tail region 408–420 was important for the CKB interaction (Fig. 2A). The C-tail amino acids 408–420 (GQLMPSKMDTCSS) partially overlap with sequences potentially important for receptor internalization (31). We also designed a series of CKB truncation mutants to locate domains important for the interaction with the PAR-1 C-tail. Interestingly, we found that by deleting the last 24 or 84 amino acids, the interaction of CKB mutants 1–357 and 1–297 with the C-tail was enhanced. The last 24–84 amino acids contain domains that distinguish CKB from other creatine kinase isoforms (32). No binding was detected with CKB mutant 1–184, which encompassed the amino-terminal region absent from the original yeast two-hybrid clone. On the other hand, the intervening CKB amino acid sequence 185–297 was sufficient for the interaction (Fig. 2B); this region is highly conserved between species and tissue isoforms and contains residues important for enzymatic catalysis (32). None of the CKB mutants bound GST alone (data not shown). Together, the binding studies revealed a specific in vitro interaction between the PAR-1 C-tail and CKB that persisted in vivo and raised the possibility that CKB may play a role in PAR-1 signaling.
Figure 2.
Domain analysis of PAR-1 association with CKB. (A) Schematic of GST–C-tail truncation mutants (Top). Pull-down assay shows 35S-labeled CKB binds to GST–C-tail mutant 382–420 (lane 3) but not to mutant 382–408 (lane 4) or 382–399 (lane 5) (Middle). Coomassie blue stain of GST and GST–C-tail fusion proteins (Bottom). Autoradiograph (AR). (B) Schematic of wild-type CKB (WT) and mutants (Upper). Autoradiograms displaying 35S-labeled CKB mutants (Left) and their binding to GST–PAR-1 C-tail fusion protein in pull-down assays (Right).
CKB Antisense Prevents Thrombin-Mediated Shape Changes in Astrocytes.
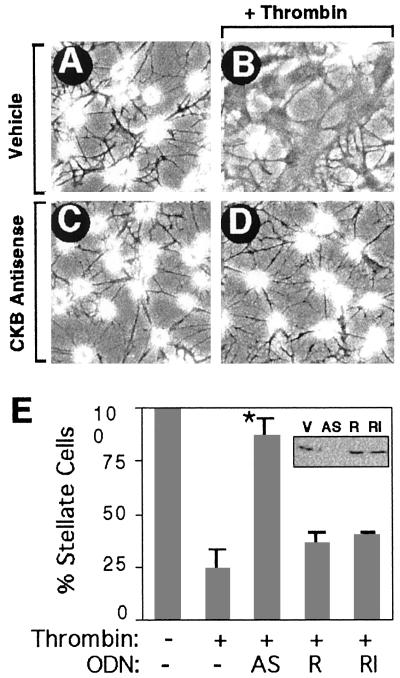
PAR-1-mediated changes in cellular structure are easily observed in the highly polarized shapes of astrocytes where cell bodies flatten and long cellular processes are resorbed (11, 33). Primary cultures of astrocytes normally display a stellate shape characterized by a round, refractile cell body with numerous processes (Fig. 3A). Upon PAR-1 activation by thrombin, most cells flattened and resorbed processes (Fig. 3B) (34). To test whether CKB was required for the cytoskeletal response, we transfected an antisense oligonucleotide (ODN) directed toward the 3′ end of the CKB mRNA to knockdown CKB expression. Similar antisense strategies were applied successfully to creatine kinase (35, 36). Astrocytes treated with CKB antisense ODNs expressed significantly lower levels of CKB detected by Western blot (Fig. 3E Inset) without affecting cell morphology (Fig. 3C). When challenged with thrombin, antisense-treated astrocytes no longer responded, and more than a 3-fold greater number of cells maintained a stellate morphology (Fig. 3D). The observed effect was not due to total ATP depletion as CKB antisense treatment did not affect total cellular ATP levels or cell viability, and no changes in actin expression levels were noted in immunoblots (data not shown). The specificity of the antisense effect was confirmed with control ODNs. The antisense ODN was modified with 2′-O-methyl groups such that it retains its ability to bind target sequences but is no longer a substrate for RNaseH, making it an extremely rigorous control for nonspecific ODN effects (37). Neither a reverse sequence ODN (Rev) nor the ODN rendered insensitive to RNaseH had any significant effect on CKB expression or on PAR-1-mediated morphological effects (Fig. 3E). An unrelated ODN with similar GC content had no effect either (data not shown). These results support a role for CKB in PAR-1-mediated cytoskeletal regulation.
Figure 3.
CKB antisense oligonucleotides prevent thrombin-induced morphological response in astrocytes. (A) Vehicle-treated cells display a stellate morphology. (B) Thrombin treatment (10 pM) converts cells to a flat, nonstellate morphology. (C) CKB antisense alone has no effect. (D) CKB antisense treatment blocks thrombin morphology effects. (E) Results were quantified as the percent stellate astrocytes, mean ± SEM., n = 3; asterisk denotes P < 0.005. (Inset) CKB immunoblot shows CKB expression levels and specificity of antisense knockdown. Vehicle (V), CKB antisense (AS), and control oligonucleotides: CKB reverse sequence (R) and CKB RNaseH-insensitive oligonucleotides (RI).
Dominant Negative CKB Blocks Shape Changes in Retinoblasts.
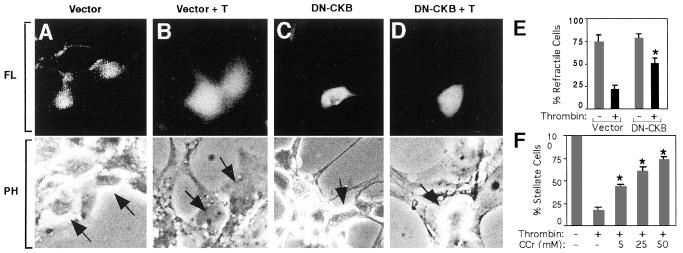
The next question we addressed was whether the catalytic activity of CKB was necessary during the PAR-1 signal. Cytosolic creatine kinase is a dimer that catalyzes the reversible transfer of phosphate from phosphocreatine to ADP in the reaction: phosphocreatine + ADP ↔ creatine + ATP (38). Expression of dominant negative CKB in which the highly conserved cysteine-283 residue near the active site is mutated to serine diminishes enzymatic activity (28). We transfected a plasmid expressing a bicistronic mRNA encoding dominant negative CKB and an enhanced green fluorescent protein reporter gene (EGFP) into human neuronal retinoblast cells (Ad12HER10) and observed the effect on PAR-1 signaling. The response to thrombin in these human neuronal cells has been extensively characterized and was especially suited for transfecting human CKB constructs (39, 40). Most cells expressing EGFP vector alone displayed round refractile cell bodies (Fig. 4A). These spherical cell bodies under phase contrast illumination appear surrounded by a bright white halo. Upon thrombin treatment, cells appeared nonrefractile, becoming flattened with increased cell body diameters (Fig. 4B). Retinoblasts expressing dominant negative CKB appeared similar to controls (Fig. 4C); these cells, however, did not flatten in response to thrombin, leaving more than a 2-fold greater number of cells with a round and refractile shape (Fig. 4D). Thus, catalytically active CKB was required to transmit the full effect of PAR-1 morphology signals. Retinoblast morphology responses were similarly inhibited by overexpression of the C-tail or C-tail peptides overlapping the CKB binding regions, whereas overexpression of CL-2 had no effect (unpublished observations).
Figure 4.
Reduction in CKB activity by dominant negative CKB (DN-CKB) expression or cyclocreatine blocks thrombin-mediated morphological changes. Transiently transfected retinoblasts were identified under fluorescence microscopy (FL) by the expression of enhanced green fluorescent protein reporter gene (Upper). Morphology of the corresponding cells (Lower; arrows) was examined under phase-contrast microscopy (PH). (A) Vector transfected cells are round and refractile. (B) Thrombin (T, 160 nM) flattens cells. (C) DN-CKB transfected cells are refractile. (D) Cells expressing DN-CKB remain refractile after thrombin treatment and do not flatten, whereas surrounding untransfected cells become flat. (E) Results were quantified as the percent transfected cells with refractile morphology, mean ± SD, n = 3; asterisk denotes P < 0.001. (F) Cyclocreatine (CCr) protects astrocytes from thrombin (10 pM) induced shape changes in a dose-dependent manner. Results are mean ± SD, n = 3; P < 0.001.
Cyclocreatine Inhibits Thrombin-Mediated Shape Changes.
The necessity for CKB enzymatic activity was further supported in experiments using the creatine analog, cyclocreatine. Although the analog is readily phosphorylated to phosphocyclocreatine, it is a very poor phosphate donor and specifically reduces ATP production by CKB (41, 42). Astrocytes treated with cyclocreatine displayed a dose-dependent decrease in their morphological response to thrombin that was phenotypically identical to the CKB antisense knockdown treatment. Although most stellate-shaped astrocytes flattened and resorbed processes in response to thrombin, up to a 4-fold greater number of astrocytes remained stellate in the presence of cyclocreatine (Fig. 4E). Cyclocreatine also blocked the thrombin/PAR-1 morphological response in Ad12HER10 retinoblasts (data not shown). Cyclocreatine had no effect on total ATP levels or cell viability (data not shown).
CKB Is Necessary for RhoA Activation But Not Calcium Signals.
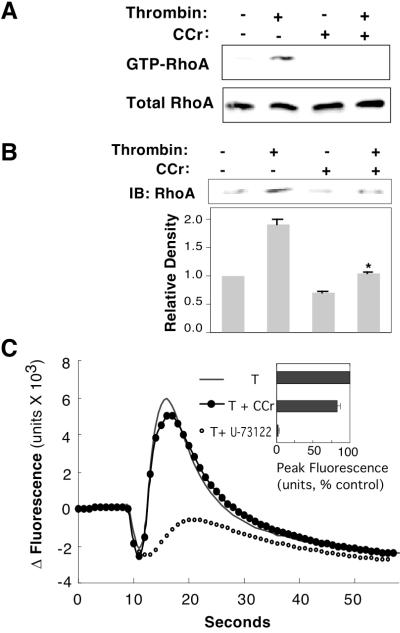
Our next step was to determine in which PAR-1 second-messenger pathways CKB might act. The Rho family of small GTP-binding proteins relays signals from PAR-1 to the actin cytoskeleton (18, 43). After PAR-1 activation, RhoA is activated with the exchange of GDP for GTP and translocates to the membrane where the sequential activation of Rho kinase, myosin light chain kinase, and myosin light chain stimulate actomyosin contraction (11, 43). To determine whether CKB activity was required in this PAR-1 pathway, we treated astrocytes with cyclocreatine and measured activated RhoA in a pull-down assay using GST–rhotekin that only interacts with activated GTP–RhoA (29). GST alone did not pull down GTP–RhoA (data not shown). The application of cyclocreatine significantly reduced GTP–RhoA after thrombin treatment (Fig. 5A). RhoA levels nearly double at the membrane after thrombin treatment (Fig. 5B). In the presence of cyclocreatine, membrane RhoA levels were similar to controls; however, there was almost no translocation after thrombin treatment (Fig. 5B). No changes in cytosolic levels of RhoA were observed, suggesting only a small fraction of RhoA translocates to the membrane in comparison to cytosolic levels (data not shown). These experiments suggest that CKB is necessary for RhoA activation and translocation to the membrane.
Figure 5.
CKB inhibition blocks PAR-1-mediated RhoA activation and translocation to the membrane but not [Ca2+]i flux. (A) Thrombin-activated GTP-RhoA (Upper) shows reduced levels with cyclocreatine treatment; RhoA levels (Lower) in total lysate (representative immunoblots, n = 3). (B) Immunoblot (IB) of RhoA in solubilized astrocyte membranes after various treatments (Upper); cyclocreatine (CCr). The relative amount of membrane RhoA was quantitated densitometrically as the percent RhoA in control cells (Lower), mean ± SD, n = 3; asterisk denotes P < 0.001. (C) CKB inhibition does not affect PAR-1-mediated increases in [Ca2+]i. Representative experiment displays the kinetics of [Ca2+]i flux monitored with the FLIPR detection system in astrocytes after thrombin addition (10 pM, T), cyclocreatine (50 mM), or phospholipase C inhibitor (1 μM, U-73122). Peak calcium changes were quantitated and normalized to control (Inset), mean ± SD, n = 3.
PAR-1 also activates phospholipase C, which leads to a rapid release of calcium from intracellular stores, and inhibition of this second messenger system has no effect on thrombin-mediated cytoskeletal reorganization (44). The calcium signal is mediated through the PAR-1 CL2 and does not require the C-tail (45). Because CKB does not bind CL2, we hypothesized that calcium signals may be independent of CKB. Intracellular calcium levels in astrocytes were monitored with the indicator dye Fluo-3. As expected, thrombin elicited a robust calcium peak observed within seconds that could be inhibited by the application of phospholipase C inhibitor (U-73122) as described (46). Application of cyclocreatine to inhibit CKB activity left the calcium response to thrombin largely intact, and cells displayed only a minor decline in peak intracellular calcium (Fig. 5C). Therefore, CKB seemed less important for the calcium second-messenger pathway.
Discussion
We identified an interaction between the PAR-1 C-tail and CKB in a yeast two-hybrid screen, and this interaction was confirmed in vitro and in vivo. Mutational studies suggested the interaction was specific and could be localized to distinct domains of CKB and the PAR-1 C-tail. Among seven-transmembrane receptors, the C-tail is one of the least conserved regions, and several mutational studies emphasize its role during receptor signaling (30, 47). Although the precise functions of PAR-1 intracellular segments are not known, the intracellular calcium release pathway was previously shown to depend on the PAR-1 CL-2 and not on its C-tail (45). The PAR-1 C-tail is required for receptor down-regulation, and recent studies demonstrate that a C-tail truncation mutant led to defects in chemotaxis (48). Thus, as with the β-adrenergic receptor and prostaglandin E receptor subtype EP3, the C-tail may help direct one of several independent signaling pathways through specific effector interactions (49, 50).
By the targeted reduction of CKB activity through three different mechanistic approaches, the efficiency of PAR-1 signals to the cytoskeletal was reduced. CKB antisense, dominant negative CKB, and competitive substrate inhibition with cyclocreatine all produced similar phenotypes. Although each method has its limitations and potential nonspecific effects, all together they support a model in which CKB is important for PAR-1 morphological signals. None of the treatments was toxic to cells nor did any treatment affect total ATP levels. Moreover, the continued presence of calcium signals after thrombin treatment strongly suggests that CKB inhibition does not disturb cell viability or thrombin signal transduction in general. This is consistent with the results of other antisense, dominant negative, knockout, and cyclocreatine studies where creatine kinase serves as a subcellular, compartmentalized regulator of ATP homeostasis rather than pancellular generator of ATP such as oxidative phosphorylation (22, 27, 38, 42).
PAR-1 calcium signals are mediated through CL2 and Gαq activation (45). This pathway is distinct from a second PAR-1 pathway mediated by Gα12/13 and RhoA that leads to actomyosin contractions underlying changes in cell morphology (51). Our studies support the concept that two separate signaling pathways emanate from PAR-1 and suggest that the C-tail may direct a Gα12/13 and RhoA pathway in which CKB activity is critical. The PAR-1–CKB interaction identified in these studies may also be important during other RhoA pathway-dependent thrombin signals that regulate cell viability, vascular endothelial permeability, platelet aggregation, and fibroblast stress fiber formation (8, 18, 43, 52). CKB is also expressed in endothelium, platelets, and fibroblasts where PAR-1 mediates morphological responses (53, 54). Preliminary studies in our laboratory suggest a similar CKB signaling mechanism may persist in these cells (unpublished observations).
Our studies suggest that PAR-1 may incorporate an ATP generating system into its signal transduction machinery by promoting a specific interaction with CKB through its C-tail. This would provide high-energy phosphate during cytoskeletal reorganization precisely at the membrane site of receptor signaling, where ATP homeostasis is critical for localized protein phosphorylation, actin polymerization, and actomyosin contraction (10, 18, 44). Mislocalization of otherwise enzymatically normal glycolytic enzymes, for example, is sufficient to disrupt normal actomyosin contractility and underscores the importance of site-specific subcellular energy generation (55). Because the dominant negative mutant is probably capable of binding to PAR-1, it is likely that localized ATP generation is a critical factor in signal generation. A role for creatine kinase during cellular shape changes and cell motility is supported by studies that showed creatine kinase-muscle knockout mice display altered contractility patterns (27). Also, creatine kinase inhibitors reduce tumor cell motility (42), and the creatine kinase homolog arginine kinase is selectively concentrated in motile versus static neuronal growth cones (26). Why certain cellular reactions are preferentially coupled to ATP generated specifically by creatine kinase as opposed to other ATP generating systems remains enigmatic, although metabolite channeling within multienzyme complexes and the energetic advantage of the close apposition of creatine kinase to ATPases demonstrated in previous studies is an attractive explanation (23, 38, 54, 56). An explicit function for creatine kinase during signal transduction is suggested by the recently demonstrated interaction with AMP-activated protein kinase and regulation of its kinase activity (57). CKB might also regulate protein kinases during PAR-1 signaling where shape changes are, in fact, inhibited by the application of drugs that block the ATP binding sites of serine-threonine kinases (10, 44), although the precise kinases for thrombin signaling are yet to be identified.
Acknowledgments
We thank P. Nothacker and O. Civelli for assistance with the calcium assays. This work was supported by National Institutes of Health Program Project Grant AG00538 (to D.D.C).
Abbreviations
- PAR-1
protease-activated receptor-1
- C-tail
cytoplasmic tail
- CL
cytoplasmic loop
- CKB
creatine kinase–brain isoform
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Grand R J, Turnell A S, Grabham P W. Biochem J. 1996;313:353–368. doi: 10.1042/bj3130353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donovan F M, Pike C J, Cotman C W, Cunningham D D. J Neurosci. 1997;17:5316–5326. doi: 10.1523/JNEUROSCI.17-14-05316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Even-Ram S, Uziely B, Cohen P, Grisaru-Granovsky S, Maoz M, Ginzburg Y, Reich R, Vlodavsky I, Bar-Shavit R. Nat Med. 1998;4:909–914. doi: 10.1038/nm0898-909. [DOI] [PubMed] [Google Scholar]
- 4.Vu T K, Hung D T, Wheaton V I, Coughlin S R. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 5.Weinstein J R, Gold S J, Cunningham D D, Gall C M. J Neurosci. 1995;15:2906–2919. doi: 10.1523/JNEUROSCI.15-04-02906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niclou S P, Suidan H S, Pavlik A, Vejsada R, Monard D. Eur J Neurosci. 1998;10:1590–1607. doi: 10.1046/j.1460-9568.1998.00183.x. [DOI] [PubMed] [Google Scholar]
- 7.Hartwig J H, Bokoch G M, Carpenter C L, Janmey P A, Taylor L A, Toker A, Stossel T P. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 8.Essler M, Amano M, Kruse H J, Kaibuchi K, Weber P C, Aepfelbacher M. J Biol Chem. 1998;273:21867–21874. doi: 10.1074/jbc.273.34.21867. [DOI] [PubMed] [Google Scholar]
- 9.Gurwitz D, Cunningham D D. Proc Natl Acad Sci USA. 1988;85:3440–3444. doi: 10.1073/pnas.85.10.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suidan H S, Stone S R, Hemmings B A, Monard D. Neuron. 1992;8:363–375. doi: 10.1016/0896-6273(92)90302-t. [DOI] [PubMed] [Google Scholar]
- 11.Suidan H S, Nobes C D, Hall A, Monard D. Glia. 1997;21:244–252. doi: 10.1002/(sici)1098-1136(199710)21:2<244::aid-glia7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Offermanns S, Mancino V, Revel J P, Simon M I. Science. 1997;275:533–536. doi: 10.1126/science.275.5299.533. [DOI] [PubMed] [Google Scholar]
- 13.Offermanns S, Laugwitz K L, Spicher K, Schultz G. Proc Natl Acad Sci USA. 1994;91:504–508. doi: 10.1073/pnas.91.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridley A J, Hall A. EMBO J. 1994;13:2600–2610. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 16.Hall A. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 17.Nobes C D, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt A, Hall M N. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 19.Jalink K, van Corven E J, Hengeveld T, Morii N, Narumiya S, Moolenaar W H. J Cell Biol. 1994;126:801–810. doi: 10.1083/jcb.126.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhanasekaran N, Dermott J M. Cell Signal. 1996;8:235–245. doi: 10.1016/0898-6568(96)00048-4. [DOI] [PubMed] [Google Scholar]
- 21.George E B, Schneider B F, Lasek R J, Katz M J. Cell Motil Cytoskeleton. 1988;9:48–59. doi: 10.1002/cm.970090106. [DOI] [PubMed] [Google Scholar]
- 22.Wallimann T, Dolder M, Schlattner U, Eder M, Hornemann T, O'Gorman E, Ruck A, Brdiczka D. Biofactors. 1998;8:229–234. doi: 10.1002/biof.5520080310. [DOI] [PubMed] [Google Scholar]
- 23.Tombes R M, Shapiro B M. Cell. 1985;41:325–334. doi: 10.1016/0092-8674(85)90085-6. [DOI] [PubMed] [Google Scholar]
- 24.Sata M, Sugiura S, Yamashita H, Momomura S, Serizawa T. Circulation. 1996;93:310–317. doi: 10.1161/01.cir.93.2.310. [DOI] [PubMed] [Google Scholar]
- 25.Friedman D L, Roberts R. J Comp Neurol. 1994;343:500–511. doi: 10.1002/cne.903430311. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y E, Esbensen P, Bentley D. J Neurosci. 1998;18:987–998. doi: 10.1523/JNEUROSCI.18-03-00987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steeghs K, Benders A, Oerlemans F, de Haan A, Heerschap A, Ruitenbeek W, Jost C, van Deursen J, Perryman B, Pette D, et al. Cell. 1997;89:93–103. doi: 10.1016/s0092-8674(00)80186-5. [DOI] [PubMed] [Google Scholar]
- 28.Lin L, Perryman M B, Friedman D, Roberts R, Ma T S. Biochim Biophys Acta. 1994;1206:97–104. doi: 10.1016/0167-4838(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 29.Ren X D, Kiosses W B, Schwartz M A. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourne H R. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro M J, Trejo J, Zeng D, Coughlin S R. J Biol Chem. 1996;271:32874–32880. doi: 10.1074/jbc.271.51.32874. [DOI] [PubMed] [Google Scholar]
- 32.Muhlebach S M, Gross M, Wirz T, Wallimann T, Perriard J C, Wyss M. Mol Cell Biochem. 1994;133:245–262. doi: 10.1007/BF01267958. [DOI] [PubMed] [Google Scholar]
- 33.Cavanaugh K, Gurwitz D, Cunningham D D, Bradshaw R. J Neurochem. 1990;54:1735–1743. doi: 10.1111/j.1471-4159.1990.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 34.Grabham P, Cunningham D D. J Neurochem. 1995;64:583–591. doi: 10.1046/j.1471-4159.1995.64020583.x. [DOI] [PubMed] [Google Scholar]
- 35.Enjolras N, Godinot C. Mol Cell Biochem. 1997;167:113–125. doi: 10.1023/b:mcbi.0000009692.67331.d3. [DOI] [PubMed] [Google Scholar]
- 36.Ch'ng J L, Mulligan R C, Schimmel P, Holmes E W. Proc Natl Acad Sci USA. 1989;86:10006–10010. doi: 10.1073/pnas.86.24.10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang M Y, Chan H, Zounes M A, Freier S M, Lima W F, Bennett C F. J Biol Chem. 1991;266:18162–18171. [PubMed] [Google Scholar]
- 38.Wallimann T, Wyss M, Brdiczka D, Nicolay K, Eppenberger H M. Biochem J. 1992;281:21–40. doi: 10.1042/bj2810021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinstein J R, Lau A L, Brass L F, Cunningham D D. J Neurochem. 1998;71:1034–1050. doi: 10.1046/j.1471-4159.1998.71031034.x. [DOI] [PubMed] [Google Scholar]
- 40.Grabham P W, Grand R J, Byrd P J, Gallimore P H. Exp Eye Res. 1988;47:123–133. doi: 10.1016/0014-4835(88)90029-2. [DOI] [PubMed] [Google Scholar]
- 41.Boehm E A, Radda G K, Tomlin H, Clark J F. Biochim Biophys Acta. 1996;1274:119–128. doi: 10.1016/0005-2728(96)00018-7. [DOI] [PubMed] [Google Scholar]
- 42.Mulvaney P T, Stracke M L, Nam S W, Woodhouse E, O'Keefe M, Clair T, Liotta L A, Khaddurah-Daouk R, Schiffmann E. Int J Cancer. 1998;78:46–52. doi: 10.1002/(sici)1097-0215(19980925)78:1<46::aid-ijc9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 43.Seasholtz T M, Majumdar M, Brown J H. Mol Pharmacol. 1999;55:949–956. doi: 10.1124/mol.55.6.949. [DOI] [PubMed] [Google Scholar]
- 44.Jalink K, Moolenaar W H. J Cell Biol. 1992;118:411–419. doi: 10.1083/jcb.118.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verrall S, Ishii M, Chen M, Wang L, Tram T, Coughlin S R. J Biol Chem. 1997;272:6898–6902. doi: 10.1074/jbc.272.11.6898. [DOI] [PubMed] [Google Scholar]
- 46.Ubl J J, Reiser G. Glia. 1997;21:361–369. doi: 10.1002/(sici)1098-1136(199712)21:4<361::aid-glia3>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 47.Gudermann T, Schoneberg T, Schultz G. Annu Rev Neurosci. 1997;20:399–427. doi: 10.1146/annurev.neuro.20.1.399. [DOI] [PubMed] [Google Scholar]
- 48.Sambrano G R, Coughlin S R. J Biol Chem. 1999;274:20178–20184. doi: 10.1074/jbc.274.29.20178. [DOI] [PubMed] [Google Scholar]
- 49.Luttrell L M, Ferguson S S, Daaka Y, Miller W E, Maudsley S, Della Rocca G J, Lin F, Kawakatsu H, Owada K, Luttrell D K, et al. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 50.Namba T, Sugimoto Y, Negishi M, Irie A, Ushikubi F, Kakizuka A, Ito S, Ichikawa A, Narumiya S. Nature (London) 1993;365:166–170. doi: 10.1038/365166a0. [DOI] [PubMed] [Google Scholar]
- 51.Klages B, Brandt U, Simon M I, Schultz G, Offermanns S. J Cell Biol. 1999;144:745–754. doi: 10.1083/jcb.144.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaughan P J, Pike C J, Cotman C W, Cunningham D D. J Neurosci. 1995;15:5389–5401. doi: 10.1523/JNEUROSCI.15-07-05389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shibata S, Kobayashi B. Thromb Res. 1975;7:417–424. doi: 10.1016/0049-3848(75)90036-5. [DOI] [PubMed] [Google Scholar]
- 54.Wallimann T, Hemmer W. Mol Cell Biochem. 1994;133–134:193–220. doi: 10.1007/BF01267955. [DOI] [PubMed] [Google Scholar]
- 55.Wojtas K, Slepecky N, von Kalm L, Sullivan D. Mol Biol Cell. 1997;8:1665–1675. doi: 10.1091/mbc.8.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srere P A. Annu Rev Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- 57.Ponticos M, Lu Q L, Morgan J E, Hardie D G, Partridge T A, Carling D. EMBO J. 1998;17:1688–1699. doi: 10.1093/emboj/17.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]