Abstract
Proteins of the ERV1/ALR family are encoded by all eukaryotes and cytoplasmic DNA viruses for which substantial sequence information is available. Nevertheless, the roles of these proteins are imprecisely known. Multiple alignments of ERV1/ALR proteins indicated an invariant C-X-X-C motif, but no similarity to the thioredoxin fold was revealed by secondary structure predictions. We chose a virus model to investigate the role of these proteins as thiol oxidoreductases. When cells were infected with a mutant vaccinia virus in which the E10R gene encoding an ERV1/ALR family protein was repressed, the disulfide bonds of three other viral proteins—namely, the L1R and F9L proteins and the G4L glutaredoxin—were completely reduced. The same outcome occurred when Cys-43 or Cys-46, the putative redox cysteines of the E10R protein, was mutated to serine. These two cysteines were disulfide bonded during a normal virus infection but not if the synthesis of other viral late proteins was inhibited or the E10R protein was expressed by itself in uninfected cells, suggesting a requirement for an upstream viral thiol oxidoreductase. Remarkably, the cysteine-containing domains of the E10R and L1R viral membrane proteins and the glutaredoxin are in the cytoplasm, in which assembly of vaccinia virions occurs, rather than in the oxidizing environment of the endoplasmic reticulum. These data indicated a viral pathway of disulfide bond formation in which the E10R protein has a central role. By extension, the ERV1/ALR family may represent a ubiquitous class of cellular thiol oxidoreductases that interact with glutaredoxins or thioredoxins.
Keywords: poxvirus, vaccinia virus, thiol oxidoreductase, glutaredoxin
Disulfide bonds, required for the stability and function of many proteins, are formed in the relatively oxidizing environment of the eukaryotic endoplasmic reticulum or the bacterial periplasm (1). Because poxviruses replicate entirely within the cytoplasm, reports of disulfide bonds in viral core proteins and the cytoplasmic domains of membrane proteins (2–5) are intriguing. Just as these large, double-stranded DNA viruses encode their own enzymes for cytoplasmic DNA and RNA synthesis (6), they also could encode proteins that participate in thiol-disulfide metabolism and protein folding. Vaccinia virus, the prototypal member of the poxvirus family, has two glutaredoxins, encoded by the O2L (7) and G4L (8) ORFs. The O2L glutaredoxin appears to be involved in nucleotide biosynthesis and is neither conserved in other poxviruses nor required for replication of vaccinia virus in cultured cells (9). In contrast, the G4L glutaredoxin is conserved in all poxviruses and recently has been shown to be cytoplasmic and required for the assembly of vaccinia virions (10). Another candidate disulfide bond-forming enzyme, the vaccinia virus E10R protein, contains the thiol active-site motif C-X-X-C (11).
The E10R protein is conserved in all poxviruses for which sequence information is available and is a member of the ERV1/ALR family, which appears to be represented in all eukaryotes (11). This large family includes the Saccharomyces cerevisiae ERV1 (Essential for Respiration and Vegatative growth) protein, which is required for mitochondrial biogenesis (12), and its homologs in other organisms, the mammalian hematopoetin (alternatively named ALR for its role as an Augmenter of Liver Regeneration) (13), and animal and plant quiescins, so called because of their up-regulation in quiescent cells (14). The precise functions of these proteins, however, remain unknown (15). The conserved domain of the ERV1/ALR family consists of ≈100 aa and contains a C-X-X-C motif, which prompted the suggestion that ERV1/ALR proteins might function as thiol oxidoreductases (11). In agreement with this, the C-X-X-C motif of the ERV1 domain is the redox-active disulfide bridge of secreted egg-white sulfhydryl oxidase, a member of the quiescin family (16).
An initial biochemical and genetic characterization of the E10R protein (33) indicated that it is (i) synthesized after viral DNA replication as a 12-kDa unprocessed polypeptide, (ii) associated with immature and mature virus particles, (iii) exposed on the membrane surface (cytoplasmic face) of purified virions, and (iv) required for virion morphogenesis. Here, we report that the cysteines of the C-X-X-C motif of the vaccinia virus E10R protein are disulfide bonded and that expression of E10R is required for the formation of the active-site disulfide bond in the G4L glutaredoxin and three pairs of disulfides in the cytoplasmic domain of the L1R viral membrane protein and a related protein encoded by the F9L ORF.
Materials and Methods
Expression Plasmids.
ORFs were amplified by PCR by using DNA from the WR strain of vaccinia virus as the template. Late vaccinia virus promoter sequences were included at the 5′ end of the forward PCR primers upstream of the ATG start codon. To express genes under the control of a T7 or cytomegalovirus (CMV) promoter, forward PCR primers starting with the ATG codon of the corresponding gene and the same reverse primers as above were used, and the PCR products were cloned in the pTargeT expression vector (Promega).
Transfection of Vaccinia Virus-Infected Cells with Recombinant Plasmids for Transient Expression.
BS-C-1 cells in a 24-well plate were infected with 10 plaque-forming units of wild-type vaccinia virus, mutant viruses, or recombinant vaccinia virus vTF7–3 expressing the T7 RNA polymerase gene (17). After 1.5 h, the infected cells were transfected with a plasmid in Lipofectamine (Life Technologies, Gaithersburg, MD) according to the manufacturer's protocol. Where indicated, cytosine arabinonucleoside (AraC) at 40 μM was added 2 h before infection and isopropyl β-d-thiogalactoside (IPTG) was added 2 h after transfection.
Western Blot Analysis.
Infected or infected and transfected BS-C-1 cells were collected 24 h after infection and washed with PBS. The alkylating agent AMS (4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid; Molecular Probes) or NEM (N-ethylmaleimide; Sigma) or the reducing agent TCEP [tris-(2-carboxyethyl)phosphine; Molecular Probes] was added to the cell pellet at a final concentration of 20 mM. The samples were dissolved in 100 mM Tris⋅HCl, pH 7.5/1% SDS. After addition of TCEP, the pH was adjusted to 7.5 with 2 M Tris, pH 8.5. The lysate was sonicated and mixed with gel-loading buffer (with or without 2-mercaptoethanol), boiled, and loaded on an SDS-polyacrylamide gel. A 16% or a 10–20% polyacrylamide gradient Tricine gel (NOVEX, San Diego) was used to resolve AMS-alkylated forms of small proteins, and a 10–20% polyacrylamide gradient Tris-glycine gel (NOVEX) was used for other separations. Proteins were transferred to nitrocellulose, incubated with a primary antibody followed by a peroxidase-conjugated secondary antibody or with a primary antibody conjugated to peroxidase, and detected with a chemiluminescent detection kit (Pierce).
Results
The E10R Protein Contains a Disulfide Bond.
Iterative database searches using the psi-blast program (18) showed that homologs of the E10R protein, comprising the ERV1/ALR family, are present in all cytoplasmic DNA viruses (other poxviruses, African swine fever virus, iridoviruses, and Paramecium bursaria Chlorella virus 1) as well as in all eukaryotes for which a significant fraction of the genome sequence is available. A multiple alignment (19) of the conserved regions of the viral and eukaryotic members of the ERV1/ALR family is shown in Fig. 1. Our suggestion (11), that these proteins comprise a structurally unique family of thiol oxidoreductases, was based on the conservation of the C-X-X-C motif, the absence of detectable sequence similarity with thioredoxin-fold proteins, the confident prediction of four α-helices (20), which rules out structural similarity to the β-sheet-based thioredoxin fold, and the inability of secondary-structure-based threading (21) to support a specific relationship with any of the known α-helical folds. We considered that a viral member of the family could provide a good model with which to investigate the role of these proteins as thiol oxidoreductases.
Figure 1.
Multiple alignment of the ERV1/ALR protein family. The protein designations consist of the Gene Identification (GI) numbers, gene names, and abbreviated species names. The positions of the aligned amino acid residues in each sequence are indicated in front of the sequences. The lengths of the poorly conserved spacers between the aligned regions are indicated. The lymphocystis disease virus (LDV) homolog of E10R and Plasmodium falciparum homolog of ERV1 are not annotated as proteins in the GenBank database and were identified by searching the corresponding genome sequences (L63545 and AL031745) using the tblastn program. The consensus derived by using the 90% conservation is shown underneath the alignment; b indicates “big” residues (E, K, R, I, L, M, F, Y, and W), h indicates hydrophobic residues (A, C, F, I, L, M, V, W, and Y), s indicates small residues (A, C, S, T, D, N, V, G, and P), and p indicates polar residues (D, E, H, K, N, Q, R, S, and T). The conserved cysteines predicted to form a redox-active center are highlighted by reverse shading. The multiple-alignment-based secondary structure prediction is shown on top of the alignment; H (h) indicates α-helix (uppercase indicates the most confident prediction). E10Rh and ERV1h refer to homologs of E10R and ERV1. Species abbreviations: VAC, vaccinia virus; RFV, rabbit fibroma virus; MYX, myxoma virus; MCV, molluscum contagiosum virus; FPV, fowlpox virus; MSV, Melanoplus sanguinipes entomopoxvirus; AFSV, African swine fever virus; LDV1, lymphocystis disease virus 1 (an iridovirus); PBCV1, Paramecium bursaria Chlorella virus 1; Sce, Saccharomyces cerevisiae, Spo, Schizosaccharomyces pombe; Hsa, Homo sapiens; Cel, Caenorhabditis elegans; Dme, Drosophila melanogaster; Lma, Leishmania major; Pfa, Plasmodium falciparum; Ath, Arabidopsis thaliana.
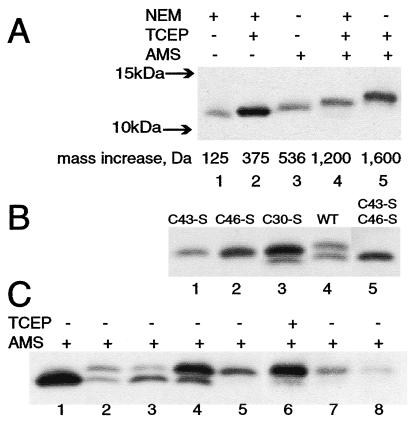
The E10R protein has three cysteines; Cys-43 and Cys-46 comprise the invariant C-X-X-C motif whereas Cys-30 is conserved in only a few closely related poxviruses (Fig. 1). Initial experiments indicated that reduced and nonreduced monomeric forms of the E10R protein have similar mobilities on SDS/PAGE. Therefore, the oxidation states of the three cysteines in the E10R protein were determined by covalent modification with the thiol-conjugating agents NEM (125 Da) or AMS (536 Da) under denaturing conditions (22). Cells were infected with a recombinant vaccinia virus vE10R-HA encoding a hemagglutinin (HA)-epitope tag at the C terminus of the E10R ORF that enabled the protein to be detected with a specific antibody. NEM or AMS was added before cell lysis with SDS to avoid oxidation during extraction. The proteins then were resolved by SDS/PAGE and detected by Western blotting. The mobility of the E10R-HA protein after alkylation of free thiol(s) with NEM or alkylation of all three cysteines by reduction with TCEP and treatment with NEM was determined. If there were one free thiol and one disulfide, the difference in mobility of the proteins in lanes 1 and 2 of Fig. 2A would be only 250 Da, consistent with the slight difference seen. As shown in lane 3 of Fig. 2A, treatment of the E10R-HA protein with AMS in the absence of TCEP decreased its mobility slightly more than adding NEM in the presence of TCEP, indicating one free thiol. Evidence for a disulfide was obtained by first treating the protein with NEM to alkylate the free thiol and then reducing the disulfide with TCEP and alkylating with AMS. These modifications should increase the mass of the E10R-HA polypeptide by 1,200 Da, consistent with the further reduction in mobility seen in lane 4 of Fig. 2A. Finally, if the E10R-HA polypeptide were reduced with TCEP and then alkylated with AMS, the net increase in mass would be 1,600 Da, resulting in a still lower mobility (Fig. 2A, lane 5). These data were consistent with the presence of one free thiol and one disulfide. Results similar to those shown in Fig. 2A were obtained when disulfide interchange was blocked by treatment of the cells with trichloroacetic acid before alkylation or when virions that had been purified from the cytoplasm of infected cells were analyzed.
Figure 2.
Thiol-disulfide state of the E10R protein. (A) Evidence for a disulfide bond. Replicate cell monolayers were infected with vE10R-HA. After 24 h, the cells were harvested and the cell pellets were treated with the following reagents in the presence of SDS: NEM (lane 1), TCEP and NEM (lane 2), AMS (lane 3), NEM followed by TCEP and then AMS (lane 4) and TCEP and AMS (lane 5). Samples were separated by SDS/PAGE, and the proteins were detected by Western blotting using a peroxidase-conjugated anti-HA antibody (clone 3F10; Roche). The predicted mass increases resulting from the addition of NEM or AMS are indicated. The arrows on the left indicate the mobilities of marker proteins. (B) Cys-43 and Cys-46 form the disulfide bond. Cells were infected with vaccinia virus and transfected with a plasmid containing the late vaccinia virus P11 promoter regulating the wild-type (WT) or a mutated E10R-HA ORF. C43-S, C46-S, and C30-S are the mutated ORFs with the respective cysteines replaced by serines. Samples were alkylated with AMS and separated by SDS/PAGE, and the proteins were detected by Western blotting by using peroxidase-conjugated anti-HA antibody. (C) Additional viral proteins are required for disulfide bond formation. BS-C-1 cells were infected with: vE10R-HA (lanes 1 and 6), wild-type vaccinia virus (lane 2), vE10Ri in absence of inducer (lane 3), vTF7–3 expressing T7 RNA polymerase (lane 4), and vTF7–3 in the presence of the DNA replication inhibitor, AraC (lane 5). The infected cells were transfected with plasmid containing E10R-HA under vaccinia virus P11 promoter (lanes 2 and 3) or T7 promoter (lanes 4 and 5) or not transfected (lanes 1 and 6). In lane 7, Cos cells were transfected with a plasmid containing the E10R-HA under the CMV promoter, and in lane 8, the E10R-HA protein was synthesized in vitro. All samples were treated with AMS alone except for the one in lane 6 that first was reduced with TCEP. Proteins were resolved by SDS/PAGE and detected by Western blotting by using a peroxidase-conjugated anti-HA antibody.
Identification of the Cysteines That Form the Disulfide Bridge of the E10R Protein.
Expression of the E10R ORF is required for replication of vaccinia virus. Previously, we showed that transfection of a plasmid containing the E10R-HA ORF regulated by the vaccinia virus P11 promoter could rescue a recombinant vaccinia virus that had its E10R gene repressed (33). Rescue also occurred when Cys-30 was changed to serine but not when either Cys-43 or Cys-46 was mutated. To determine the effects of these mutations on disulfide bond formation, we infected cells with wild-type vaccinia virus and transfected them with plasmids containing a wild-type or mutated E10R-HA ORF regulated by the P11 promoter. The cells were lysed in the presence of AMS, and the E10R-HA protein was detected by SDS/PAGE and Western blotting. Although nearly all of the E10R-HA protein was in the disulfide-bonded form when expressed from a recombinant vaccinia virus (Fig. 2A), only about half was disulfide-bonded when expressed by transfection. This was evident from the detection of two bands with the mobilities expected for modification of one cysteine (lower band) and three cysteines (upper band) with AMS (Fig. 2B, lane 4). Two bands also were detected when Cys-30 was mutated (Fig. 2B, lane 3), but, here, they had mobilities consistent with two and no cysteines modified by AMS, suggesting that Cys-43 and Cys-46 formed the disulfide bond. Further support for this was obtained by mutating either Cys-43 or Cys-46. In both conditions, only a single band with two cysteines modified by AMS was detected because of the absence of a disulfide bond (Fig. 2B, lanes 1 and 2). When both Cys-43 and Cys-46 were mutated, a single band with one cysteine modified by AMS was resolved (Fig. 2B, lane 5). These results demonstrated that Cys-43 and Cys-46 of the E10R protein form an intramolecular disulfide bond.
E10R Disulfide Bond Formation Is Dependent on Synthesis of Additional Viral Proteins.
In the experiments described above, the E10R-HA ORF was regulated by a vaccinia virus late promoter and transfected into cells that had been productively infected with wild-type vaccinia virus. To determine whether the formation of the E10R disulfide bond was dependent on other viral proteins, we placed the E10R-HA ORF under control of the CMV promoter and transfected the plasmid into uninfected cells. As mobility markers, some cells were infected with vE10R-HA and treated with AMS to modify the single free cysteine (Fig. 2C, lane 1) or reduced with TCEP and treated with AMS so all three cysteines would be modified (Fig. 2C, lane 6). When E10R-HA under the CMV promoter was transfected into uninfected cells, all three cysteines of the E10R-HA protein reacted with AMS (Fig. 2C, lane 7), indicating the absence of a disulfide bond. The above experiment suggested that one or more vaccinia virus proteins were needed for the formation of disulfide-bonded E10R.
Further experiments were designed to determine whether the viral proteins needed for the oxidation of the E10R protein belonged to the pre- or postreplicative class. Before vaccinia virus DNA replication or when DNA replication is inhibited, only early-stage genes are expressed, whereas after DNA replication both intermediate- and late-stage genes are turned on (6). Because the E10R gene belongs to the late class, it would not be expressed in the presence of a DNA synthesis inhibitor. To circumvent this, we infected cells with a recombinant vaccinia virus, vTF7–3, that expresses T7 RNA polymerase even in the presence of an inhibitor of DNA replication. A plasmid with the E10R-HA ORF regulated by a T7 promoter then was transfected into these infected cells. In the absence of inhibitor, both disulfide-bonded and reduced forms of the E10R-HA protein were seen (Fig. 2C, lane 4) whereas in the presence of the inhibitor only the reduced form was detected (Fig. 2C, lane 5). This result suggested that the viral proteins required for the oxidation of the E10R protein (as E10R itself) belong to the postreplicative class.
We noted that when the E10R-HA protein was expressed by using the powerful T7 expression system, the ratio of disulfide bonded to reduced forms was lower (Fig. 2C, lane 4) than when the vaccinia virus P11 promoter was used (Fig. 2C, lane 2). This suggested that the relative amount of disulfide-bonded E10R protein might vary inversely with the total amount of E10R protein, which comprised both the untagged and HA-tagged species encoded by the virus and the plasmid, respectively. To investigate this, we infected cells with the mutant virus vE10Ri, which has an inducible E10R gene, in the absence of IPTG and transfected them with a plasmid containing the E10R-HA ORF regulated by the vaccinia virus P11 promoter. The ratio of disulfide bonded to reduced E10R-HA was greater when expression of the viral E10R gene was repressed (Fig. 2C, lane 3) than when it was active (Fig. 2C, lane 2). We also found that the E10R-HA was entirely disulfide-bonded when the relatively weak, natural E10R promoter was used to express the transfected E10R-HA gene (data not shown). The dependence of the relative amounts of the reduced and oxidized forms of E10R on the expression level resembles the pattern seen for the bacterial periplasmic thiol oxidoreductase DsbA (23).
Influence of the E10R Protein on the Thiol-Disulfide State of the Vaccinia Virus G4L Protein.
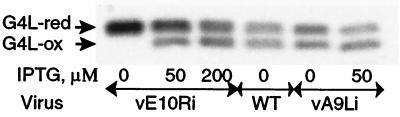
G4L is a glutaredoxin homolog, with two cysteine residues in a C-X-X-C motif, whose redox activity has been demonstrated in vitro (8). Additional studies indicated that G4L is present in the cytoplasm of infected cells and packaged in virus particles (10). Because the mobilities of the reduced and unreduced G4L proteins were similar when analyzed by SDS/PAGE, we again used thiol alkylation with AMS to impart size differences. Analysis of G4L protein from wild-type vaccinia virus-infected cells that were lysed in the presence of AMS indicated that about half of the molecules contained a disulfide bond and migrated faster than the reduced and alkylated form (Fig. 3). A similar result was obtained when the cells were infected with vE10Ri, an inducible E10R mutant, in the presence of 200 μM IPTG, although the ratio of disulfide-bonded to reduced forms was slightly less when 50 μM IPTG was used. In the absence of IPTG, the disulfide-bonded form of the G4L glutaredoxin was not detected, indicating a requirement for expression of E10R. This was not due to the block in morphogenesis that results from repression of E10R because repression of the A9L gene of the inducible mutant vA9Li, which also interrupts morphogenesis at a similar stage (24), did not prevent oxidation of the G4L protein (Fig. 3).
Figure 3.
E10R expression is required for the disulfide bond in the G4L protein. Cells were infected with vE10Ri or vA9Li or wild-type (WT) vaccinia virus in the presence or absence of 50 or 200 μM IPTG. Samples were alkylated with AMS and separated by SDS/PAGE, and the proteins were detected by Western blotting by using polyclonal anti-G4L antibody (10) followed by peroxidase-conjugated anti-HA antibody. Arrows indicate the reduced (red) and disulfide-bonded (ox) forms of the G4L protein.
Influence of the E10R Protein on the Thiol-Disulfide State of Vaccinia Virus L1R and F9L Proteins.
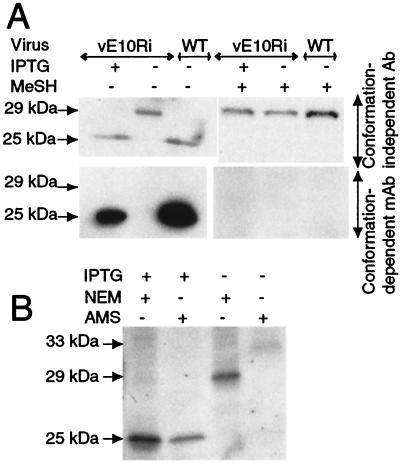
The protein encoded by the L1R ORF of vaccinia virus has six cysteines that are conserved in orthologs from all poxviruses (11). The L1R protein is a myristoylated, disulfide-bonded component of intracellular mature enveloped virions, with most of the protein, including the portion containing the cysteines, exposed on the cytoplasmic face of the virion (5, 25). Previous studies indicated that the unreduced and reduced forms of the L1R protein are conformationally distinct. This difference was revealed by the more rapid migration of unreduced L1R compared with reduced L1R on SDS/PAGE and the inability of certain mAbs to react with the reduced form of the protein (5, 26). Using a polyclonal antibody (27), we found that the L1R protein existed in the fast-migrating, disulfide-bonded state when expressed in cells infected with wild-type virus or vE10Ri in the presence of IPTG (Fig. 4A). In contrast, the L1R protein expressed by vE10Ri in the absence of IPTG had the slow mobility of the fully reduced form (Fig. 4A). Moreover, the L1R protein synthesized under the latter condition did not react with a conformation-dependent mAb whereas the protein did react this way when cells were infected with vE10Ri in the presence of IPTG or with wild-type virus (Fig. 4A). Thus, the formation of L1R disulfide bonds depended on expression of the E10R protein.
Figure 4.
E10R expression is required for disulfide bonds in the L1R protein. (A) BS-C-1 cells were infected with the vE10Ri in the presence (+) or absence (−) of IPTG or with wild-type (WT) vaccinia virus. The cells were lysed with SDS in the presence of NEM and analyzed by PAGE with (+) or without (−) prior reduction with mercaptoethanol (MeSH). The L1R protein was detected by Western blotting by using either conformation-independent polyclonal L1R antibody (Upper) or conformation-dependent anti-L1R mAb (Lower) followed by peroxidase-conjugated antibody. (B) Cells were infected in the presence or absence of IPTG as in A, lysed in the presence (+) of NEM or AMS, and analyzed by SDS/PAGE without prior reduction as in A. The estimated masses (kDa) of different forms of the L1R protein are shown on the left.
The L1R protein contains six cysteine residues, but the number that form disulfide pairs had not been determined. To obtain this information, we infected cells with vE10Ri in the presence of IPTG and lysed them in the presence of NEM or AMS. The mobility of the L1R protein on SDS/PAGE was similar with either alkylating agent (Fig. 4B), suggesting that all six cysteines were disulfide-bonded and, therefore, unreactive. In contrast, when cells were infected with vE10Ri in the absence of IPTG, the L1R protein migrated more slowly when treated with AMS than with NEM (Fig. 4B). The difference in mobilities between the NEM- and AMS-treated L1R protein was approximately 3 kDa, equivalent to six AMS residues, suggesting that all six cysteines were in the reduced thiol form (Fig. 4B). Thus, formation of the three disulfide pairs of the L1R protein was dependent on coexpression of E10R.
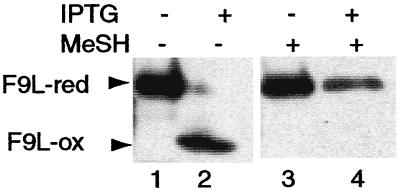
The F9L protein was studied next because it has the same predicted membrane topology and pattern of six invariant cysteines as the L1R protein. Cells were infected with vE10Ri in the presence or absence of IPTG and transfected with a plasmid encoding a HA-tagged F9L ORF regulated by a vaccinia virus strong synthetic late promoter. The cells were lysed in the presence of NEM and analyzed by SDS/PAGE with or without reduction by mercaptoethanol. In the presence of IPTG, the unreduced form of the F9L protein had a more rapid mobility than the reduced form, indicating the presence of disulfide bonds (Fig. 5). In contrast, the F9L protein was in the reduced form when E10R was not expressed (Fig. 5). Thus, disulfide bond formation of the two homologous proteins, L1R and F9L, required expression of E10R.
Figure 5.
E10R expression is required for disulfide bonds in the F9L protein. Cells were infected with the vE10Ri in the presence or absence of IPTG and transfected with a plasmid encoding the F9L ORF with a C-terminal HA-tag regulated by a modified, synthetic, strong late promoter. Samples were lysed in the presence of NEM or reduced with mercaptoethanol, resolved by SDS/PAGE with or without prior reduction, and analyzed by Western blotting by using peroxidase-conjugated anti-HA antibody. Arrows on the left point to the reduced (red) and disulfide-bonded (ox) forms of the F9L protein.
Both Cysteines of the C-X-X-C Motif Are Critical for the Role of the E10R Protein in Disulfide-Bond Formation.
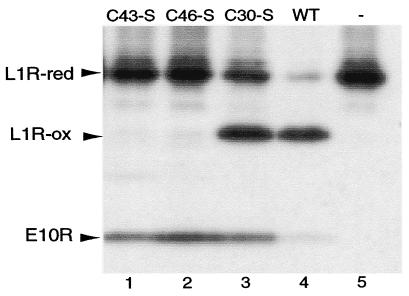
We assessed the effect of each of the three Cys-Ser substitutions of the E10R protein on L1R and F9L disulfide bond formation by cotransfecting the L1R-HA plasmid with plasmids expressing mutated or wild-type E10R into cells infected with vE10Ri in the absence of IPTG. The rapidly migrating disulfide-bonded form of L1R-HA protein was detected only when the unmutated E10R or the Cys-30 → Ser mutated form was transfected (Fig. 6). The absence of disulfide-bonded L1R protein when the Cys-43 → Ser or the Cys-46 → Ser mutated gene was transfected was not a result of poor expression or stability of the E10R-HA protein as shown by Western blotting (Fig. 6). We did notice, however, a greater amount of reduced L1R when the Cys-30 → Ser mutated form was transfected compared with the wild type. Results similar to those obtained by transfecting the L1R-HA ORF were obtained by transfecting the F9L-HA ORF (data not shown). These data demonstrated that an intact C-X-X-C motif was essential for the role of the E10R protein in disulfide bond formation whereas the third, nonconserved cysteine of the E10R protein was not required.
Figure 6.
Effect of cysteine mutations in the E10R protein on the formation of disulfide bonds in the L1R protein. Cells were infected with vE10Ri, transfected with a plasmid containing the P11 promoter regulating the L1R ORF with a C-terminal HA tag, and cotransfected with a plasmid having the P11 promoter regulating the E10R-HA ORF with all three cysteines or with a single cysteine mutated to serine. Samples were alkylated with NEM, resolved by SDS/PAGE, and detected by Western blotting using peroxidase-conjugated anti-HA antibody. The reduced (red) and disulfide-bonded (ox) forms of the L1R protein are indicated by arrows. After alkylation with NEM, the reduced and disulfide-bonded forms of the E10R protein are not resolved from each other.
Discussion
The presence of disulfide bonds in the cytoplasmic domains of vaccinia virus proteins led to the suggestion that vaccinia virus may have acquired a unique pathway of disulfide bond formation (4, 11). Here, we provide evidence for such a pathway involving virus-encoded proteins. The E10R protein, a member of the ERV1/ALR family of eukaryotic and viral proteins, was shown to have a disulfide bond between Cys-43 and Cys-46 and to directly or indirectly mediate disulfide bond formation of at least three other viral proteins. In the absence of E10R expression, the two cysteines of the G4L glutaredoxin and the six cysteines of the L1R and F9L proteins remained in the reduced state. Mutation of Cys-43 or Cys-46 but not Cys-30 to serine also abrogated disulfide bond formation of the L1R and F9L proteins. Studies of the E10R protein suggested that the redox-active disulfide bond is located on the outer or cytoplasmic face of the membrane of intracellular mature virions (33), compatible with its interaction with the cytoplasmic domains of the G4L, L1R, and F9L proteins. Oxidation of the E10R protein did not occur in uninfected cells that were transfected with a plasmid containing the E10R ORF controlled by a CMV promoter or in vaccinia virus-infected cells in the presence of an inhibitor of DNA replication, suggesting that at least one other late viral protein functions in the disulfide bond formation pathway upstream of E10R. The G4L glutaredoxin may act as an intermediate between the E10R protein and the downstream L1R and F9L proteins because E10R was disulfide-bonded and L1R and F9L were reduced when G4L expression was repressed (T.G.S. and C.L.W., unpublished data).
Given the role that E10R has in the formation of disulfide bonds in virion proteins, it is not surprising that repression of E10R expression would result in an arrest of morphogenesis (33). The opposite possibility, however, that the defect in morphogenesis was responsible for the failure of disulfide bond formation, also was considered. That this was not the case was shown by studies with inducible A9L (24) and A17L (28) mutants. We found that disulfide bond formation of G4L, L1R, and F9L proteins was unaffected when expression of the A9L membrane protein was repressed, even though morphogenesis was blocked at a stage similar to that occurring with the E10R mutant (Fig. 3 and data not shown). Moreover, disulfide bond formation also occurred when assembly was blocked at an even earlier stage by repression of the A17L membrane protein or by addition of the drug rifampicin (data not shown). Thus, the impairment of disulfide bond formation appeared to be caused directly by the E10R mutation and is probably responsible for inhibition of morphogenesis. Locker and Griffiths (4) reported that vaccinia virus morphogenesis was only partially impaired when infected cells were treated with DTT, but the oxidation states of the E10R, G4L, L1R, and F9L proteins were not among those determined, and, therefore, they might not have been completely reduced.
Cytoplasmic disulfide bond formation is not unprecedented, because thioredoxins and glutaredoxins undergo cycles of oxidation and reduction of cysteine pairs as part of their enzymatic activity (29). What is unusual is the presence of cytoplasmic proteins with stable disulfide bonds. However, Escherichia coli mutants in which the reduction of thioredoxin and glutathione is impaired accumulate disulfide-bonded proteins in the cytoplasm (30, 31). The cytoplasmic factory regions in which vaccinia virus assembly occurs might exclude cellular disulfide reductases and, thus, provide a local environment conducive to disulfide bond formation catalyzed by virus-encoded enzymes.
The demonstration of the role of E10R, a ERV1/ALR family protein, in disulfide bond formation is of general interest because it provides a clue as to the possible biological function of this highly conserved but poorly characterized eukaryotic protein family. Our finding that E10R is required for the oxidation of G4L, a glutaredoxin, implies a functional interaction between these two unrelated proteins in a disulfide bond formation pathway. This makes an interesting parallel to the fusion of ERV and thioredoxin domains in quiescins (14, 16). Perhaps all ERV1/ALR family proteins function as thiol oxidoreductases in conjunction with thioredoxins or glutaredoxins. This hypothesis is compatible with the conservation of E10R homologs in all cytoplasmic DNA viruses, each of which also encodes at least one glutaredoxin or thioredoxin, and is conceptually similar to the recently described pathway of disulfide bond formation in the endoplasmic reticulum in which the ERO1p protein acts upstream of the thioredoxin-domain protein disulfide isomerase (32). In the present model, ERV1/ALR proteins would functionally replace the structurally unrelated ERO1p.
Acknowledgments
D. Hruby and Y. Ichihashi generously donated antibodies to the L1R protein, and W. Yeh kindly provided recombinant vaccinia virus vA9Li. We thank L. Aravind for helpful discussions and G. Storz for comments on the manuscript.
Abbreviations
- CMV
cytomegalovirus
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- HA
influenza virus hemagglutinin
- E10R-HA
vaccinia virus E10R ORF with C-terminal HA epitope
- IPTG
isopropyl β-d-thiogalactoside
- NEM
N-ethylmaleimide
- TCEP
tris-(2-carboxyethyl)phosphine
- vE10Ri
recombinant vaccinia virus with inducible E10R gene
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210397997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210397997
References
- 1.Rietsch A, Beckwith J. Annu Rev Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- 2.Ichihashi Y. Virology. 1981;113:277–284. doi: 10.1016/0042-6822(81)90154-9. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez J F, Paez E, Esteban M. J Virol. 1987;61:395–404. doi: 10.1128/jvi.61.2.395-404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locker J K, Griffiths G. J Cell Biol. 1999;144:267–279. doi: 10.1083/jcb.144.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolffe E J, Vijaya S, Moss B. Virology. 1995;211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]
- 6.Moss B. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 2637–2671. [Google Scholar]
- 7.Ahn B Y, Moss B. Proc Natl Acad Sci USA. 1992;89:7060–7064. doi: 10.1073/pnas.89.15.7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gvakharia B O, Koonin E, Mathews C K. Virology. 1996;226:408–411. doi: 10.1006/viro.1996.0669. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopal I, Ahn B Y, Moss B, Mathews C K. J Biol Chem. 1995;270:27415–27418. doi: 10.1074/jbc.270.46.27415. [DOI] [PubMed] [Google Scholar]
- 10.White C L, Weisberg A S, Moss B. J Virol. 2000;74:9175–9183. doi: 10.1128/jvi.74.19.9175-9183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 12.Lisowsky T. Curr Genet. 1994;26:15–20. doi: 10.1007/BF00326299. [DOI] [PubMed] [Google Scholar]
- 13.Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter K A, Starzl T. Proc Natl Acad Sci USA. 1994;91:8142–8146. doi: 10.1073/pnas.91.17.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppock D L, Cina-Poppe D, Gilleran S. Genomics. 1998;54:460–468. doi: 10.1006/geno.1998.5605. [DOI] [PubMed] [Google Scholar]
- 15.Polimeno L, Lisowsky T, Francavilla A. Ital J Gastroenterol Hepatol. 1999;31:494–500. [PubMed] [Google Scholar]
- 16.Hoober K L, Glynn N M, Burnside J, Coppock D L, Thorpe C. J Biol Chem. 1999;274:31759–31762. doi: 10.1074/jbc.274.45.31759. [DOI] [PubMed] [Google Scholar]
- 17.Fuerst T R, Niles E G, Studier F W, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rost B, Sander C, Schneider R. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 21.Rost B, Schneider R, Sander C. J Mol Biol. 1997;270:471–480. doi: 10.1006/jmbi.1997.1101. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. Proc Natl Acad Sci USA. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishigami S, Akiyama Y, Ito K. FEBS Lett. 1995;364:55–58. doi: 10.1016/0014-5793(95)00354-c. [DOI] [PubMed] [Google Scholar]
- 24.Yeh, W. W., Moss, B. & Wolffe, E. J. (2000) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 25.Ravanello M P, Hruby D E. J Gen Virol. 1994;75:1479–1483. doi: 10.1099/0022-1317-75-6-1479. [DOI] [PubMed] [Google Scholar]
- 26.Ichihashi Y, Oie M. Virology. 1996;220:491–494. doi: 10.1006/viro.1996.0337. [DOI] [PubMed] [Google Scholar]
- 27.Ravanello M P, Franke C A, Hruby D E. J Biol Chem. 1993;268:7585–7593. [PubMed] [Google Scholar]
- 28.Wolffe E J, Moore D M, Peters P J, Moss B. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 30.Derman A I, Prinz W A, Belin D, Beckwith J. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 31.Bessette P H, Aslund F, Beckwith J, Georgiou G. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frand A R, Kaiser C A. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 33.Senkevich, T. G., Weisberg, A. S. & Moss, B. (2000) Virology, in press. [DOI] [PubMed]