Abstract
Dbl-homology guanine nucleotide exchange factors (DH-GEFs) regulate actin cytoskeletal reorganization, cell adhesion, and gene transcription via activation of Rho GTPases. However, little is known about the physiological role of mammalian DH-GEFs during development. The DH-GEF family member Trio is of particular interest because it is a multifunctional protein possessing two GEF domains, as well as a protein serine/threonine kinase domain, and trio-like genes in Caenorhabditis elegans and Drosophila were shown to function in neural migration and axon guidance. To determine the role of Trio during mammalian development, we generated a mouse trio loss-of-function mutation (trio−/−). Trio function is essential during late embryonic development as genotype analysis indicated that trio−/− embryos died between embryonic day (E)-15.5 and birth, or shortly thereafter. In the trio−/− embryos, primary skeletal myofibers were relatively normal at E14.5, but by E18.5 highly unusual spherical myofibers accumulated. Trio deficiency may cause a defect in secondary myogenesis, as the appearance of the abnormal trio−/− skeletal myofibers temporally coincided with the onset of secondary myogenesis, and smaller secondary myofibers located adjacent to the primary myofibers were absent. The proliferation of trio−/− secondary myoblasts appeared normal, suggesting that Trio may regulate secondary myoblast alignment or fusion. trio−/− embryos also displayed aberrant organization in several regions within the brain, including the hippocampal formation and olfactory bulb. We thus conclude that Trio is essential for late embryonic development, and that Trio functions in fetal skeletal muscle formation and in the organization of neural tissues.
Keywords: gene targeting, embryonic lethality, Rac, mouse development
Rho family GTPases are key regulators of actin cytoskeletal reorganization, cell adhesion, and gene transcription (1–3). These GTPases are activated by Dbl-homolgy guanine nucleotide exchange factors (DH-GEFs), which promote the exchange of GDP for GTP (4, 5). In the GTP-bound activated state, Rho family GTPases interact with diverse effector targets such as protein kinases, lipid kinases, and structural proteins (2, 4–9). However, little is known about the physiological function of mammalian DH-GEF genes during development, with the exception of the mouse vav gene and the human faciogenital dysplasia (FGD1) gene (10–12). Among the about two dozen DH-GEFs, Trio is unusual as it contains two GEF domains, with each domain having specificity for different Rho family GTPases both in vitro and in vivo (13–16). Furthermore, Trio is a large and complex protein which also contains pleckstrin homology (PH) domains, multiple spectrin-like repeats, two SH3 domains, an Ig-like domain, and a kinase domain that is most closely related to calcium/calmodulin dependant kinases (13). Trio was originally isolated as a binding protein to LAR transmembrane protein tyrosine phosphatase and it was suggested that a Trio–LAR complex might integrate diverse signals needed to coordinate actin cytoskeleton remodeling during cell migration and growth (13). Ectopic expression studies indicate a role for Trio in regulating actin cytoskeletal reorganization, cell migration, and cell growth (15).
Trio belongs to a subfamily of DH-GEFs, which includes Duet (17), Kalirin (18–19), Caenorhabditis elegans UNC-73 (20), and Drosophila Trio (21–24). UNC-73 is necessary for both cell migration and axon guidance (20); Drosophila Trio regulates axon guidance (21–24); and Kalirin may regulate vesicle trafficking (25). Drosophila Trio has been functionally linked to Rac, p21-activated kinase (Pak), Abl protein tyrosine kinase, DLAR transmembrane protein tyrosine phosphatase, and the adaptor proteins Dock and Ena (21–24). To determine the role of mammalian Trio during development, we generated a mouse trio loss-of-function mutation (trio−/−).
Here we demonstrate that absence of Trio causes embryonic lethality associated with abnormal development of skeletal muscle and neural tissues. In the trio−/− embryos, primary skeletal myofibers were relatively normal at embryonic day (E)-14.5, but by E18.5 highly unusual spherical myofibers accumulated. Proliferation of secondary myoblasts appeared normal in trio−/− embryos, indicating that Trio may regulate secondary myoblast localization or fusion. Normal myofiber formation occurs in two stages, with primary myofibers being formed by the alignment and fusion of primary myoblasts (26, 27). Primary myofibers then serve as scaffolds for the proliferation, alignment, and fusion of secondary myoblasts into secondary myofibers beginning at about E15 of mouse development. trio−/− mouse embryos also displayed aberrant organization in several regions within the brain, suggesting that Trio, like UNC-73 and Drosophila Trio, functions in neural positioning.
Materials and Methods
Gene Targeting.
The Trio targeting vector consists of a 4.8-kb HindIII–HindIII genomic trio DNA fragment, which contains exons encoding a portion of the amino-terminal GEF domain followed by a phosphoglycerate kinase (PGK)-promoter neor cassette, a 4.5-kb HindIII–HindIII genomic trio DNA fragment containing additional exons encoding the amino-terminal GEF domain, and a PGK-thymidine kinase cassette inserted into pSP73 (see Fig. 1A). Embryonic stem (ES) J1 cells were electroporated with the NotI linearized targeting construct and selected with G418 (150 μg/ml) and ganciclovir (2 μM) (Calbiochem), and expanded for Southern analysis. Three resulting clones of ≈900 clones analyzed yielded the expected size fragments and were used for injection into BALB/c blastocysts. Blastocysts were transferred to the uterus of pseudopregnant Swiss Webster mice. Germ-line transmission of one of the clones was obtained on further crossing of male chimeras with BALB/c females.
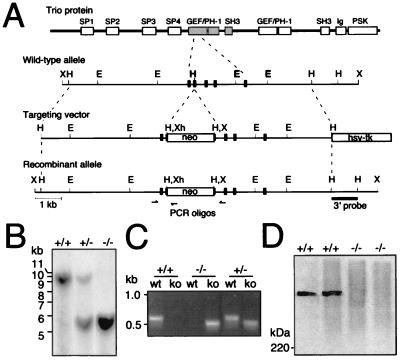
Figure 1.
Gene targeting at the trio locus. (A) The structure of the Trio protein is schematically shown at the Top. Below the structure is schematically shown a portion of the trio gene exon–intron organization with the location of restriction enzyme sites, the structure of the targeting vector, and the structure of the trio locus after integration of the targeting vector. Correct gene targeting results in an insertion of the marker gene for positive selection (PGKneo) into an exon that encodes a part of the Trio amino-terminal GEF domain. The hsv-tk (thymidine kinase) expression cassette was used for negative selection. The 3′ probe used for Southern blot analysis is shown as a solid bar, and the relative location of the PCR oligonucleotides by arrows. E, EcoRI; H, HindIII; X, XbaI; Xh, XhoI. (B) Southern blot analysis of DNA from offspring derived from heterozygous matings. Genomic DNA isolated from E14.5 embryos was digested with XbaI and analyzed by using the 3′ probe, yielding the 9.3-kb and 5.3-kb fragments expected for the wild-type allele and mutant allele, respectively. Southern blot analysis of genomic DNA using a neo probe also showed proper targeting and single insertion of the transfected vector (data not shown). +, wild-type trio allele; −, mutant trio allele. (C) PCR analysis of genomic DNA using PCR primer sets specific for the wild-type (wt) genomic configuration or for the knockout (ko) rearranged configuration. (D) Immunoblot of anti-Trio mAb immunoprecipitates from trio+/+, and trio−/− E14.5 primary cell cultures confirming the absence of Trio protein in trio−/− cells. The position of the 220-kDa size standard is shown at the left.
Genotype Analysis.
Tail biopsies or embryos were subjected to proteinase K digestion for 10–16 h at 55°C in buffer containing 50 mM Tris⋅HCl (pH 8.5), 20 mM EDTA, 10 mM NaCl, 0.5% SDS, and 0.5 mg/ml proteinase K. Genomic DNA was recovered by phenol extraction and ethanol precipitation, and ≈20 μg of genomic DNA was digested with XbaI and then used for Southern blot analysis. trio genotype analysis at E14.5 demonstrated an essentially normal distribution (trio+/+/trio+/−/trio−/− = 21:34:15), indicating no essential function for Trio through E14.5. However, at E15.5–E18.5 there was a 40–60% decrease in the expected number of trio−/− embryos, indicating that ≈60% of the trio−/− embryos die between E14.5 and E18.5 (trio+/+/trio+/−/ trio−/− at E15.5–E16.5 = 19:36:10; and at E18.5 = 27:42:8).
Protein Analysis.
For protein analysis, trio−/− and trio+/+ E14.5 embryonic cell cultures were lysed in a buffer containing 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. Insoluble material was then removed by centrifugation (10 min at 10,000 × g). Immunoprecipitations were performed by using anti-Trio mAbs essentially as previously described (13). Protein was then resolved by SDS/6% PAGE under reducing conditions, transferred to nitrocellulose membrane (Schleicher & Schuell), and blotted with an anti-Trio mAbs. Antibody was detected with protein A/G-horseradish peroxidase (Pierce) and the chemiluminescence reagent, luminol (DuPont/NEN).
Histology and Immunohistochemistry.
For histology, embryos were fixed in 10% Bouin's fixation fluid (VWR Scientific), embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) according to standard procedures. For immunohistochemical analysis, sections were deparaffinized, rehydrated, and permeabilized with 0.5% Nonidet P-40 in PBS for 5 min at ≈20°C, and then washed in PBS. For anti-BrdUrd mAb (Boehringer Mannheim) and anti-laminin mAb (Sigma) stainings, sections were initially treated with 2 M HCl for 30 min at 37°C and then 0.1 M borax for 10 min at ≈20°C. Sections used for anti-BrdUrd and anti-laminin mAb staining, as well as for antiskeletal myosin mAb (Innovex Biosciences) and anti-laminin mAb staining, were then treated with 3% H2O2 for 5 min at ≈20°C, and finally with PBS containing 10% goat serum for 30 min at ≈20°C. Sections were then incubated with the appropriate combination of mAbs for 16 h at 4°C, washed in PBS, then further incubated for 3 h at ≈20°C with Texas red-linked goat anti-mouse IgG1 (Southern Biotechnology Associates) and FITC-linked donkey anti-rabbit IgG (Jackson ImmunoResearch) diluted in PBS containing 10% (vol/vol) goat serum. For staining of sections with rabbit anti-calretinin sera (Chemicon), sections were treated in the following manner: incubated with 0.001% (wt/vol) Pronase (Calbiochem) in PBS for 30 min at ≈20°C, incubated with PBS containing 2% (vol/vol) goat serum 1 h at ≈20°C, incubated with sera diluted in PBS containing 2% goat serum for ≈16 h at 4°C, washed with PBS, incubated for 3 h at ≈20°C with horseradish peroxidase (HRP)-linked donkey anti-rabbit Ig sera (Amersham), and washed in PBS, and then HRP activity was detected by using the brown chromogen diaminobenzidine. Pregnancies were timed by housing appropriate mice overnight and then separating female mice on finding vaginal plugs, which was considered E0.5. For BrdUrd analysis, E15.5 pregnant females were given two i.p. injections of 10 mg of BrdUrd per ml at 75 μg/g body weight 8 h apart.
Results
Trio Deficiency Causes Embryonic Lethality.
The structure of Trio, containing two DH-GEF domains, each with adjacent pleckstrin homology (PH) and SH3 domains, as well as a protein serine/threonine kinase domain with an adjacent Ig-like domain, and multiple spectrin-like domains is shown schematically in Fig. 1A. To determine Trio function during development, we generated Trio-deficient mice by using ES cells containing a targeted trio mutation (the targeting vector and the wild-type and recombinant trio alleles are schematically shown in Fig. 1A). Proper integration of the Trio disruption construct into an exon encoding part of the amino-terminal GEF domain was confirmed by Southern blot analysis of mutant ES cells (data not shown). Transmission of the mutated trio allele (trio−) through the germ line was demonstrated by Southern blot (Fig. 1B) and PCR analysis (Fig. 1C) of DNA from embryos and offspring of mice derived from the trio+/− ES cells. The absence of Trio protein in trio−/− embryonic cells was confirmed by anti-Trio immunoblot analysis (Fig. 1D). Breeding of trio+/− mice demonstrated an essential role for Trio during development, as no viable trio−/− offspring were obtained (trio+/+/trio+/−/trio−/− = 74:92:0 at 3 weeks post birth). trio genotype analysis at various embryonic stages indicated that ≈60% of trio−/− embryos died between E15.5 and E18.5, and that the remaining trio−/− embryos died between E18.5 and birth, or shortly thereafter.
Trio Is Required for Normal Skeletal Muscle Development.
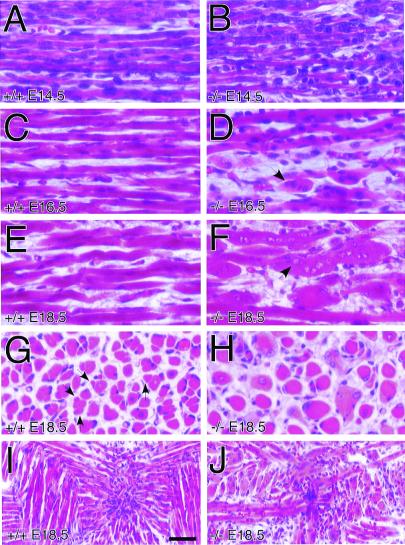
The most readily observed phenotype in the trio−/− embryos was abnormal skeletal muscle histology of limb, trunk, and head. This defect was clearly visible in sagittal sections of thoracic, paraspinal skeletal muscle in E14.5, E16.5, and E18.5 trio−/− compared with matched littermate trio+/+ embryos (Fig. 2). In the E14.5 trio−/− embryos, the primary skeletal myofibers appeared relatively normal, but were less uniformly organized than the trio+/+ fibers (Fig. 2 A and B). At E16.5, and especially by E18.5, however, trio−/− embryos contained strikingly deformed spherical skeletal myofibers (Fig. 2 C–F), and lacked smaller secondary myofibers normally adjacent to the primary myofibers (Fig. 2 G and H). The spherical shape was confirmed by analysis of serial sections of trio−/− myofibers (data not shown). The deformed muscle fibers in both the E16.5 and E18.5 embryos were multinucleated, but, whereas the E16.5 nuclei had normal H&E staining, E18.5 nuclei stained poorly, indicating normal myoblast proliferation, but progressive degeneration of nuclei (arrows in Fig. 2 D and F). Furthermore, analysis of transverse sections of E18.5 muscle groups confirmed the highly unusual trio−/− muscle morphology and showed that, whereas control embryos contained organized clusters of myofibers, trio−/− muscle lacked such organization (Fig. 2 G and H). Overall, the trio−/− myofibers were thicker and there was a general absence of secondary fibers (some secondary fibers of trio+/+ muscle, which are distinguishable by their smaller diameter in transverse sections, are marked with arrows in Fig. 2G). The skeletal muscle abnormality was also evident in coronal sections of trio−/− tongue muscle (Fig. 2 I and J). The development of smooth and cardiac muscle appeared normal (data not shown), indicating that Trio function is essential only to skeletal muscle development.
Figure 2.
Deformed trio−/− embryonic skeletal myofibers. Shown are H&E-stained sagittal sections of matched thoracic paraspinal skeletal muscle from trio+/+ (A, C, and E) and trio−/− (B, D, and F) embryos at E14.5 (A and B), E16.5 (C and D), and E18.5 (E and F) of development. From E14.5 to E18.5 there is an increase in the number of abnormal large rounded myofibers (see arrows). (G and H) H&E staining of E18.5 trio+/+ (G) and trio−/− (H) coronal sections of the same muscles shown in A–F, demonstrating the abnormally large and morphologically unusual trio−/− myofibers. Arrows in G indicate the location of some secondary myofibers in trio+/+ embryos. (I and J) H&E staining of E18.5 trio+/+ (I) and trio−/− (J) coronal sections of tongue muscle. (Scale bar represents 25 μm for panels A–H, and 50 μm for I and J.)
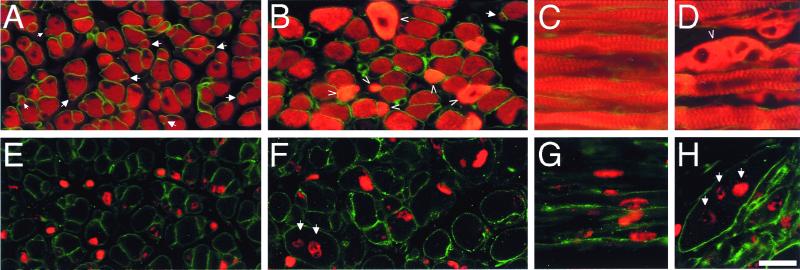
The deficiency of secondary myofibers in trio−/− embryos was confirmed by skeletal muscle myosin (red) and laminin (green) staining of E18.5 embryos (Fig. 3 A and B). Secondary myofibers, which are formed by the fusion of secondary myoblasts aligned beneath the laminin-rich basal lamina of the primary myofibers (26, 28), were largely absent in trio−/− embryos compared with trio+/+ embryos (arrows in Fig. 3 A and B). However, the lack of secondary fibers in trio−/− embryos was not due to an absence of secondary myoblast proliferation, as in utero BrdUrd labeling of E15.5 proliferating cells demonstrated that E18.5 trio−/− skeletal myofibers contained secondary myoblasts [sections were stained for BrdUrd (red) and laminin (green) as shown in Fig. 3 E and F]. Similar analysis of longitudinal sections also indicated that the deformed myofibers contained BrdUrd-labeled secondary myoblasts (arrows in Fig. 3 C, D, G, and H).
Figure 3.
Abnormal secondary skeletal muscle myogenesis in trio−/− embryos. Shown are immunofluorescence images of E18.5 trio+/+ (A, C, E, and G) and trio−/− (B, D, F, and H) skeletal myofibers stained for skeletal muscle myosin (red) and laminin (green) (A–D), or for BrdUrd (red) and laminin (green) (E–H). Proliferating cells were labeled with BrdUrd at E15.5. Compared with control skeletal myofibers, the trio−/− muscle contains secondary myofibers that appear normal (normal secondary fibers are visualized as thinner myosin staining fibers adjacent to, or beneath, the lamina of primary fibers (see arrows in A). Some trio−/− myofibers contain abnormally high levels of myosin (see arrowheads in B and D), possibly because of the abnormal fusion of secondary myoblasts or the collapse of the myofiber from its normal elongated fibrous form into a condensed spherical form. E18.5 myofibers contained E15.5 BrdUrd-labeled myoblasts (see arrows in F and H), indicating that secondary myoblast proliferation was normal at least through E15.5. trio−/− embryos also contained some striated myofibers that appeared relatively normal (D). For BrdUrd labeling, pregnant mice were injected i.p. with BrdUrd at E15.5 and embryos were sectioned at E18.5. (Scale bar represents 100 μm for A–H.)
Trio Functions in Embryonic Neural Organization.
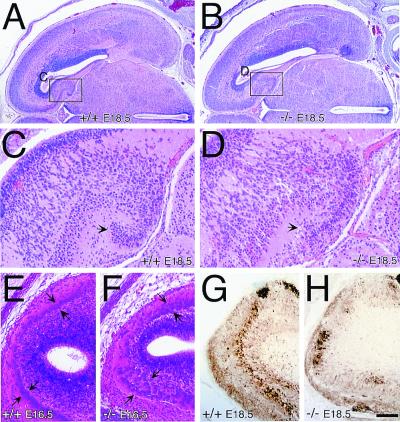
In addition to the skeletal muscle abnormality, trio−/− embryos displayed abnormal neural organization. Gross overall brain histology of trio−/− embryos appeared normal (coronal sections of E18.5 embryos are shown in Fig. 4 A and B). However, localized areas of cellular disorganization were evident within the trio−/− brains. Within the trio−/− hippocampal formation, the dentate gyrus cells displayed altered organization (arrows in Fig. 4 C and D). And within E16.5 trio−/− olfactory bulbs, the mitral cell layer was diffusely organized compared with the uniform mitral layer seen in controls (arrows in Fig. 4 E and F). Abnormal mitral cell organization was also observed in E18.5 trio−/− olfactory bulbs stained for expression of the calcium-binding protein calretinin (Fig. 4 G and H). Thus, Trio appears necessary for fine-tuning the positioning of neurons, but is not essential for overall brain development, as trio−/− embryo brains display relatively normal gross histology.
Figure 4.
Abnormal neural organization in trio−/− embryos. (A and B) H&E-stained coronal sections of trio+/+ (A) and trio−/− (B) E18.5 embryos showing normal overall gross morphology of the trio−/− brains, which display differences in localized cellular organization. (C and D) Higher magnification images of the trio+/+ (C) and trio−/− (D) hippocampal regions boxed in A and B. Arrows point to a portion of the dentate gyrus that is abnormally organized in the trio−/− embryos. (E and F) H&E-stained sagittal sections of trio+/+ (E) and trio−/− (F) E16.5 olfactory bulbs reveal abnormal organization of the mitral cell layer (arrows) in the trio−/− embryos. (G and H) Anti-calretinin-stained E18.5 sagittal sections from trio+/+ (G) and trio−/− (H) embryos. Calretinin-expressing cells, visualized by using the brown chromogen diaminobenzidine, are poorly organized in the trio−/− E18.5 embryos. The abnormal neural organization was observed in several independent trio−/− embryos, and was seen in multiple sections of individual embryos (data not shown). (Scale bar represents 420 μm for A and B, 95 μm for C and D, and 100 μm for E–H.)
Discussion
We demonstrate that the Trio DH-GEF plays an essential function during embryonic development and that trio−/− embryonic lethality is associated with abnormal skeletal muscle and neural tissue development. The muscle and neural trio−/− phenotypes appear unique, but both defects are consistent with loss of Trio-mediated activation of Rho family GTPases, as Rho GTPases were shown to play key roles in both myogenesis and neural development. For instance, both dominant-negative and constitutively active forms of Drosophila Rac1 (Drac1) cause defects in myoblast fusion (29), and dominant-negative Drac1 causes defects in motor axon guidance (30). Rho GTPases were also shown necessary for transcription of muscle-specific genes by regulating the expression of the myogenic transcription factors myogenin and MEF2 (31). Moreover, Drosophila trio was shown to interact genetically with rac1 to regulate photoreceptor and motor guidance (21, 22). Abnormal muscle patterning was also observed in the Drosophila trio mutants (22).
Whether the skeletal muscle and neural defects are the basis for the trio−/− embryonic lethality, or whether other defects cause or contribute to lethality, is unknown. The trio−/− muscle defect in itself is, however, probably not the sole cause of embryonic death, because mice deficient for the myogenin transcription factor have severely reduced amounts of skeletal muscle, but survive fetal development and die immediately after birth (32, 33). Because Trio is expressed in skeletal muscle and brain (13, 21–24), the trio−/− skeletal muscle and neural defects are probably cell-autonomous, but it is possible that Trio deficiency may affect the function of other cells necessary for skeletal muscle and neural development.
The skeletal muscle abnormality seen in the trio−/− embryos is most likely caused by a defect in secondary myogenesis because (i) E14.5 trio−/− myofibers appear normal, indicating no defect in primary myogenesis; (ii) the appearance of abnormal trio−/− skeletal myofibers temporally coincides with the onset of secondary myogenesis at about E15; and (iii) the absence of smaller secondary myofibers located adjacent to the primary myofibers in the E16.5 and E18.5 trio−/− embryos indicates a defect in secondary myofiber formation (26, 27). Furthermore, the incorporation of secondary myoblasts into the abnormal spherical trio−/− myofibers indicates that Trio is not necessary for secondary myoblast proliferation, but rather necessary for the localized migration, alignment, or fusion of secondary myoblasts to form secondary myofibers. The aberrant spherical trio−/− myofibers may arise from abnormal secondary myoblast fusion, and the thicker trio−/− primary fibers may form by improper fusion of secondary myoblasts with the primary fibers (34).
trio−/− E18.5 embryos also display aberrant organization in several regions within the brain, including the hippocampal formation and olfactory bulb. This aberrant neural organization suggests that Trio, like UNC-73 (20) and Drosophila Trio (21–24), regulates Rac-mediated actin cytoskeleton rearrangement necessary for neural organization. Such a function is consistent with previous ectopic expression studies in nonneural cells, which demonstrated that the amino-terminal Trio GEF domain promotes actin cytoskeletal rearrangement and cell migration (15). However, given that trio−/− fetal brains displayed relatively normal gross histology, Trio may only be required for fine-tuning the positioning of a subset of neurons. Indeed, comparison of the trio−/− neural phenotype with the unc-73 or Drosophila trio mutant neural phenotypes suggests that mammalian Trio may have a more restricted or specialized function in neuronal migration and axon guidance compared with UNC-73 or Drosophila Trio. Mammalian Trio function may also be partially compensated for by other DH-GEF family members, such as Kalirin (35). Because muscle development is partially dependent on innervation (36–38), it is also possible that improper innervation contributes to the trio−/− muscle deformity.
Precisely how Trio-mediated activation of Rho GTPases functions during neural and skeletal muscle development remains to be established, but with the recent findings that Drosophila Trio function is linked to p21-activated kinase (Pak), Abl, DLAR, and the adaptor proteins Dock and Ena (21–24), it is possible that mammalian Trio-mediated signaling also involves some of these proteins. Trio was isolated as a LAR binding protein (13), and a functional link was established between Drosophila Trio and DLAR in motor axon guidance (22); it is thus tempting to speculate that LAR family protein tyrosine phosphatase-mediated signaling may partially regulate Trio. Possibly, a Trio–LAR complex regulates some aspect of cell adhesion and axon cytoskeletal reorganization necessary for cell fusion and axon guidance. Why Trio deficiency affects fetal myofiber formation and neuronal organization is also unknown, but it is possible that the organization of secondary myoblasts and neurons, such as the dentate gyrus cells which proliferate and migrate relatively late during embryonic development, is particularly dependent on Trio. Trio may function in the final stages of cell migration to fine-tune the organization or alignment of cells, or to control axon guidance and muscle innervation. We conclude that Trio is essential for late embryonic development, and that Trio functions in fetal skeletal muscle formation and in the organization of neural tissues.
Acknowledgments
We thank Nancy Kedersha and Haruo Saito for critical review of the manuscript; Andreas Beck, Glenn Dranoff, Karl Herrup, Hart Lidov, Joanna Panni, Arlene Sharpe, Stuart F. Schlossman, and Li-Huei Tsai for advice; and Sherri Witt for expert technical assistance. This work was supported by grants from the National Institutes of Health to M.S. M.S. is a Scholar of the Leukemia and Lymphoma Society.
Abbreviations
- DH-GEF
Dbl-homology guanine nucleotide exchange factors
- E
embryonic day
- ES
embryonic stem
- H&E
hematoxylin and eosin
References
- 1.Mackay D J, Hall A. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- 2.Van Aelst L, D'Souza-Schorey C. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 3.Fukata M, Nakagawa M, Kuroda S, Kaibuchi K. J Cell Sci. 1999;112:4491–4500. doi: 10.1242/jcs.112.24.4491. [DOI] [PubMed] [Google Scholar]
- 4.Cerione R A, Zheng Y. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 5.Zohn I M, Campbell S L, Khosravi-Far R, Rossman K L, Der C J. Oncogene. 1998;17:1415–1438. doi: 10.1038/sj.onc.1202181. [DOI] [PubMed] [Google Scholar]
- 6.Aspenstrom P. Curr Opin Cell Biol. 1999;11:95–102. doi: 10.1016/s0955-0674(99)80011-8. [DOI] [PubMed] [Google Scholar]
- 7.Schoenwaelder S M, Burridge K. Curr Opin Cell Biol. 1999;11:274–286. doi: 10.1016/s0955-0674(99)80037-4. [DOI] [PubMed] [Google Scholar]
- 8.Lim L, Manser E, Leung T, Hall C. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 9.Chernoff J. Nat Cell Biol. 1999;1:E115–E117. doi: 10.1038/12942. [DOI] [PubMed] [Google Scholar]
- 10.Zmuidzinas A, Fischer K D, Lira S A, Forrester L, Bryant S, Bernstein A, Barbacid M. EMBO J. 1995;14:1–11. doi: 10.1002/j.1460-2075.1995.tb06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasteris N G, Cadle A, Logie L J, Porteous M E, Schwartz C E, Stevenson R E, Glover T W, Wilroy R S, Gorski J L. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 12.Olson M F, Pasteris N G, Gorski J L, Hall A. Curr Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 13.Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park S H, Streuli M. Proc Natl Acad Sci USA. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellanger J M, Lazaro J B, Diriong S, Fernandez A, Lamb N, Debant A. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- 15.Seipel K, Medley Q G, Kedersha N L, Zhang X A, O'Brien S P, Serra-Pages C, Hemler M E, Streuli M. J Cell Sci. 1999;112:1825–1834. doi: 10.1242/jcs.112.12.1825. [DOI] [PubMed] [Google Scholar]
- 16.Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouviere C, Fort P. J Cell Sci. 2000;113:729–739. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- 17.Kawai T, Sanjo H, Akira S. Gene. 1999;227:249–255. doi: 10.1016/s0378-1119(98)00605-2. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R C, Penzes P, Eipper B A, Mains R E. J Biol Chem. 2000;275:19324–19333. doi: 10.1074/jbc.M000676200. [DOI] [PubMed] [Google Scholar]
- 19.Colomer V, Engelender S, Sharp A H, Duan K, Cooper J K, Lanahan A, Lyford G, Worley P, Ross C A. Hum Mol Genet. 1997;6:1519–1525. doi: 10.1093/hmg/6.9.1519. [DOI] [PubMed] [Google Scholar]
- 20.Steven R, Kubiseski T J, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue C W, Pawson T, Culotti J. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- 21.Newsome T P, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson B J. Cell. 2000;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- 22.Bateman J, Shu H, Van Vactor D. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 23.Liebl E C, Forsthoefel D J, Franco L S, Sample S H, Hess J E, Cowger J A, Chandler M P, Shupert A M, Seeger M A. Neuron. 2000;26:107–118. doi: 10.1016/s0896-6273(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 24.Awasaki T, Saito M, Sone M, Suzuki E, Ito K, Hama C. Neuron. 2000;26:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 25.Mains R E, Alam M R, Johnson R C, Darlington D N, Back N, Hand T A, Eipper B A. J Biol Chem. 1999;274:2929–2937. doi: 10.1074/jbc.274.5.2929. [DOI] [PubMed] [Google Scholar]
- 26.Wigmore P M, Dunglison G F. Int J Dev Biol. 1998;42:117–125. [PubMed] [Google Scholar]
- 27.Gullberg D, Velling T, Lohikangas L, Tiger C F. Front Biosci. 1998;3:D1039–D1050. doi: 10.2741/a344. [DOI] [PubMed] [Google Scholar]
- 28.Maina F, Casagranda F, Audero E, Simeone A, Comoglio P M, Klein R, Ponzetto C. Cell. 1996;87:531–542. doi: 10.1016/s0092-8674(00)81372-0. [DOI] [PubMed] [Google Scholar]
- 29.Luo L, Liao Y J, Jan L Y, Jan Y N. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 30.Kaufmann N, Wills Z P, Van Vactor D. Development (Cambridge, UK) 1998;125:453–461. doi: 10.1242/dev.125.3.453. [DOI] [PubMed] [Google Scholar]
- 31.Takano H, Komuro I, Oka T, Shiojima I, Hiroi Y, Mizuno T, Yazaki Y. Mol Cell Biol. 1998;18:1580–1589. doi: 10.1128/mcb.18.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasty P, Bradley A, Morris J H, Edmondson D G, Venuti J M, Olson E N, Klein W H. Nature (London) 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 33.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Nature (London) 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M, McLennan I S. Dev Dyn. 1995;204:168–177. doi: 10.1002/aja.1002040207. [DOI] [PubMed] [Google Scholar]
- 35.Penzes P, Johnson R C, Alam M R, Kambampati V, Mains R E, Eipper B A. J Biol Chem. 2000;275:6395–6403. doi: 10.1074/jbc.275.9.6395. [DOI] [PubMed] [Google Scholar]
- 36.Ashby P R, Pincon-Raymond M, Harris A J. Dev Biol. 1993;156:529–536. doi: 10.1006/dbio.1993.1099. [DOI] [PubMed] [Google Scholar]
- 37.Ashby P R, Wilson S J, Harris A J. Dev Biol. 1993;156:519–528. doi: 10.1006/dbio.1993.1098. [DOI] [PubMed] [Google Scholar]
- 38.Washabaugh C H, Ontell M P, Shan Z, Hoffman E P, Ontell M. Dev Dyn. 1998;211:177–190. doi: 10.1002/(SICI)1097-0177(199802)211:2<177::AID-AJA6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]