Abstract
Mutational inactivation of the adenomatous polyposis coli (APC) tumor suppressor initiates most hereditary and sporadic colon carcinomas. Although APC protein is located in both the cytoplasm and the nucleus, the protein domains required to maintain a predominantly cytoplasmic localization are unknown. Here, we demonstrate that nuclear export of APC is mediated by two intrinsic, leucine-rich, nuclear export signals (NESs) located near the amino terminus. Each NES was able to induce the nuclear export of a fused carrier protein. Both APC NESs were independently able to interact with the Crm1 nuclear export factor and substitute for the HIV-1 Rev NES to mediate nuclear mRNA export. Both APC NESs functioned within the context of APC sequence: an amino-terminal APC peptide containing both NESs interacted with Crm1 and showed nuclear export in a heterokaryon nucleocytoplasmic shuttling assay. Also, mutation of both APC NESs resulted in the nuclear accumulation of the full-length, ∼320-kDa APC protein, further establishing that the two intrinsic APC NESs are necessary for APC protein nuclear export. Moreover, endogenous APC accumulated in the nucleus of cells treated with the Crm1-specific nuclear export inhibitor leptomycin B. Together, these data indicate that APC is a nucleocytoplasmic shuttle protein whose predominantly cytoplasmic localization requires NES function and suggests that APC may be important for signaling between the nuclear and cytoplasmic compartments of epithelial cells.
Mutational inactivation of the adenomatous polyposis coli (APC) tumor suppressor gene initiates the vast majority of colorectal carcinomas through the development of the adenomatous polyp, now recognized as a common precursor of colon cancer (1). These inactivating mutations span the amino-terminal half of the gene and typically result in production of a truncated APC protein. Although APC protein must ordinarily function to regulate colonocyte growth and differentiation, as indicated by the formation of adenomas in its absence, we do not yet fully understand how truncating APC mutations predispose a cell to polyp development and colorectal carcinogenesis.
An intensely examined function of the APC protein is regulation of cytoplasmic β-catenin levels through targeting of cytoplasmic β-catenin for proteolysis (2–4). In addition to its role linking E-cadherin and actin at adherens junctions, β-catenin participates in Wnt signaling. The Wnt pathway can play an important role in neoplastic transformation of mammalian cells (5, 6), as indicated by the findings that mutations in the β-catenin gene that prevent APC-mediated phosphorylation and proteolytic degradation (7, 8) are often found in colon and other tumors. These observations indicate that β-catenin regulation is important in APC tumor suppressor function.
Using immunofluorescence microscopy and biochemical fractionation, we have previously shown that full-length wild-type APC protein localizes to both the cytoplasm and the nucleus of human epithelial cells (9). Others have confirmed the presence of APC protein in the nucleus of murine intestinal cells, Xenopus A6 cells, and Drosophila epithelial cells (10–12). However, in many cells, APC appears to be predominantly cytoplasmic, with a concentration near the leading edge (9, 13). Proteins with such complex cellular distribution patterns have the potential to transmit signals between the cytoplasmic and nuclear compartments. Given APC protein's participation in the Wnt/β-catenin-mediated signaling pathway, we sought to characterize whether APC has the capacity to convey information between the nucleus and the cytoplasm.
Here, we demonstrate nuclear export of APC protein via two intrinsic nuclear export signals (NESs) and nuclear-cytoplasmic shuttling of APC protein through association with the export receptor Crm1. Demonstration that APC protein shuttles between compartments via inherent nuclear import and export signals indicates that this tumor suppressor protein might participate in signal transduction between these two compartments.
Materials and Methods
Tissue Culture.
All mammalian cell lines were maintained in 37°C incubators with 5% CO2. Cos7, 293T, 3T3, and HeLa cells were grown in DMEM with 10% FBS, and cells from the mouse fibroblast cell line REF52 were cultured as previously described (14).
Molecular Clones.
Glutathione S-transferase (GST)/peptide fusion expression constructs were made by cloning APC sequences into the bacterial expression vector pGEX-2T128/129, which contains a FLAG epitope tag in the peptide linker (14). For fusions of GST with putative APC NES (NES1APC or NES2APC), DNA oligonucleotides were synthesized to encode the appropriate NES with EcoRI overhangs on both ends and cloned into pGEX-2T128/129. Mutant NESs, containing the Leu → Ala mutations underlined in Fig. 1A, were generated in either NES1APC or in both NES1APC and NES2APC in an expression construct containing a Flag epitope tag fused to the 5′-end of full-length APC cDNA, using a PCR mutagenesis strategy (15).
Figure 1.
Two NES sequences in APC protein. (A) Alignment of the two putative NES sequences of APC protein with previously characterized leucine-rich NES of HIV-1 Rev, TFIIIA, PKI-α, and mitogen-activated protein kinase kinase. Boxed amino acids indicate important hydrophobic residues (leucine or isoleucine). Underlined amino acids are critical leucine residues, which are changed to alanine in mutant forms of NES1 and NES2. (B) Conservation of the first NES sequence of APC protein among different species. The second NES is identical among species.
The following mammalian expression plasmids have been previously described: the parental expression plasmid pBC12/cytomegalovirus (CMV); plasmids expressing the wild-type or M32 mutant form of HIV-1 Rev; plasmids expressing a Gal4/Crm1 or a Crm1/Tat fusion protein; the indicator plasmids pDM128/CMV, pSLIIB/CAT, and pG6(−31)HIVLTRΔTAR (16–18). pBC12/CMV-based plasmids expressing fusions of the Rev RNA binding domain to candidate wild-type or mutant NES1APC or NES2APC were constructed, as previously described (19). Synthetic oligonucleotides, bearing cohesive BglII and XhoI ends, that encode wild-type or mutant forms of the candidate APC NES1 (68-LLERLKELNLD-78) or NES2 (162-LQNLTKRIDSLPLT-175) were inserted into pcRevM9, 3′ to the Rev cDNA. The NES1APC and NES2APC mutants contain alanine in place of the underlined leucine residues. A pBC12/CMV-based construct encoding APC residues 1–270, either wild-type or containing both NESAPC mutations, fused to the herpes simplex virus VP16 activation domain and the simian virus 40 (SV40) T antigen nuclear localization signal, was constructed as previously described (16) after PCR amplification of the wild-type or mutant APC sequence from an APC expression plasmid.
Purification and Injection of APC Fusion Proteins.
Injection of purified GST fusion proteins into REF52 cells and subsequent immunofluorescence microscopy were performed as previously described (14).
Cellular Localization of APC Protein Following Leptomycin B Treatment.
HeLa cells were treated with 10 μg/ml leptomycin B (a generous gift from Minoru Yoshida) for 2 h and then fractionated as described (9). Equivalent amounts of cytosolic and nuclear fractions were separated by SDS/PAGE and analyzed by Western immunoblot with APC antibody Ab-1 (1:150, Oncogene Science). Relative purity of the fractions was confirmed by probing for the nuclear marker lamin (1:10, American Research, Beltsville, MD) and the cytoplasmic marker tubulin (1:200, ICN). Following development with ECL substrate (Amersham Pharmacia), images were captured using a Lumi-Imager, and bands were quantitated using Lumi-Analyst software (Boehringer Mannheim). A standard Student's t test was performed using Graph Pad Prism (San Diego, CA) for data analysis.
Localization of APC Protein Expressed in Cos7 Cells Following Transient Transfection.
Plasmid DNA was used for transfection with Qiagen Superfect (Boehringer Mannheim) or Fugene 6 (Life Technologies) as instructed by the manufacturer. Cells were fixed 24 h posttransfection and stained for APC protein using anti-Flag antibody (1:32,000, Sigma) as described (9). Nuclei were visualized with 4′,6-diamidino-2-phenylindole stain for immunofluorescence microscopy or TO-PRO3 (Molecular Probes) for confocal microscopy. Confocal microscopy was performed with an Olympus Fluoview scanning laser biological inverted microscope, IX70. For protein localization studies, 100 transfected cells for each condition were scored for Flag-APC. Results were analyzed as the mean ± SD from three independent experiments.
Rev Activity Assay.
Rev activity was measured using the previously described pDM128/CMV indicator construct (17, 20), which bears the chloramphenicol acetyltransferase (CAT) indicator gene and the HIV-1 Rev response element sequestered between functional 5′ and 3′ splice sites. Human 293T cells were transfected with pDM128/CMV and the relevant wild-type or mutant Rev expression plasmid, and induced CAT activity was measured at ≈44 h after transfection.
Two-Hybrid Interaction Assays.
The pSLIIB/CAT indicator plasmid contains an HIV-1 long-terminal repeat promoter in which the TAR element, the normal RNA target for the HIV-1 Tat transcription factor, has been replaced with the SLIIB Rev RNA binding site (16). As previously shown, coexpression of a Crm1/Tat fusion protein with wild-type Rev, but not with a Rev NES mutant, results in the activation of pSLIIB/CAT-dependent CAT expression due to the recruitment of Tat to SLIIB by the Crm1–Rev NES interaction (16). To test whether other candidate NES sequences can also bind Crm1, 293T cells were transfected with pSLIIB/CAT, pCrm1/Tat, and wild-type or chimeric Rev expression plasmids, and CAT activity was determined at ≈44 h posttransfection.
An alternative two-hybrid assay used the CAT-based indicator plasmid, pG6(−31)HIVLTRΔTAR, that contains reiterated GAL4 DNA binding sites 5′ to a minimal promoter element (18). Protein interactions are measured by coexpression of a fusion protein bearing the GAL4 DNA binding domain, in this case GAL4/Crm1, and a second fusion protein bearing the VP16 activation domain and the SV40 T antigen nuclear localization signal (NLS), in this case containing wild-type or mutant forms of APC residues 1–270. These plasmids were introduced into 293T cells by transfection, and induced CAT activities were measured ≈44 h later (16).
Heterokaryon Nucleocytoplasmic Shuttling Assay.
The heterokaryon assay was carried out as previously described (21) with the following exceptions. After fixation and permeabilization (19), the APC fusion proteins were visualized in the heterokaryons by indirect immunofluorescence using a monoclonal mouse anti-VP16 antibody (Santa Cruz Biotechnology) and rhodamine-conjugated goat anti-mouse antibody (Cappel). Figures were photographed using a Leica DMRB fluorescence microscope at a 100× magnification.
Results
APC Protein Contains Two Functional Nuclear Export Signals.
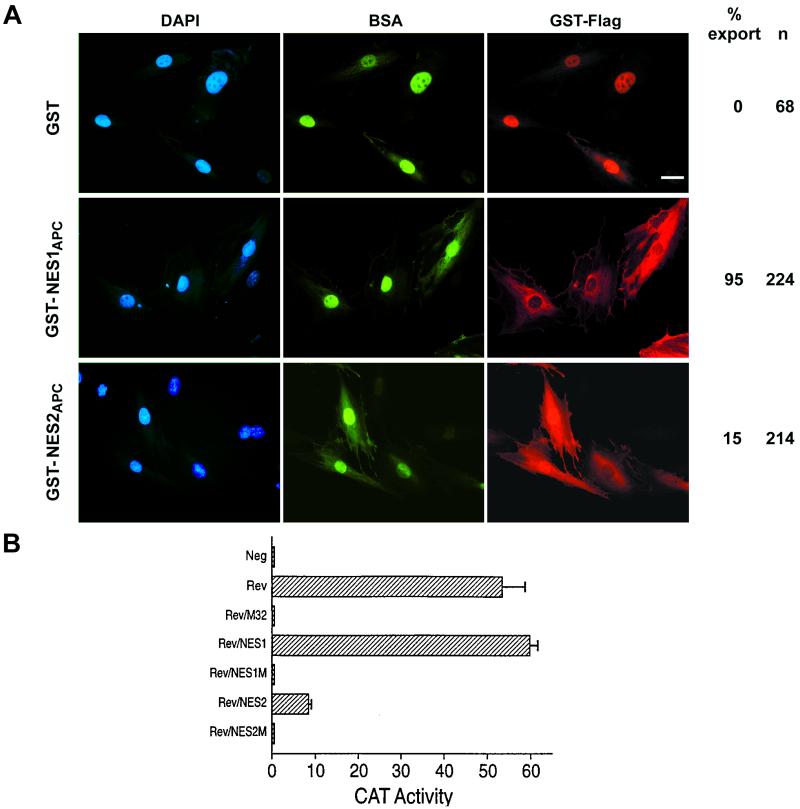
Analysis of the amino acid sequence of APC revealed two potential leucine-rich nuclear export signals, one located between amino acids 68–77 (NES1APC) and the other between amino acids 165–174 (NES2APC) (Fig. 1A). The NES1APC and NES2APC sequences are conserved among APC proteins from human, mouse, rat, and frog (Fig. 1B). To determine whether NES1APC or NES2APC was able to direct nuclear export, GST-Flag–NESAPC fusion proteins were microinjected into the nucleus of rat fibroblast cells and localized by immunofluorescence (Fig. 2A, right panels). Injected cells that displayed only cytoplasmic staining with a Flag antibody, and only nuclear staining of the BSA control protein, indicate that the test substrate has been exported from the nucleus.
Figure 2.
Both APC NESs are functional. (A) APC NESs can direct nuclear export of a linked protein. GST was purified from Escherichia coli either unmodified or fused with NES1APC or NES2APC. Proteins were injected along with FITC-BSA into the nuclei of REF52 cells. Cells were incubated for 15–30 min at 37°C to allow for nuclear export and then fixed and stained for immunofluorescence microscopy as follows: nuclei, 4′,6-diamidino-2-phenylindole (DAPI) counterstain (blue), FITC-BSA injection control (green), and Flag-tagged GST fusion protein (red). Nuclear export, defined as cells with only cytoplasmic GST-Flag staining and only nuclear BSA control staining, is shown to the right as a percentage of the cells scored. (Scale bar, 20 μm.) (B) NES1APC and NES2APC can each functionally substitute for the HIV-1 Rev NES. NES1APC or NES2APC, but not mutant (M) forms of these NESs, are able to substitute for the HIV Rev NES in mediating the nuclear export and expression of the CAT mRNA expressed by the pDM128/CMV indicator construct. Results represent the average of three independent experiments with standard deviation as indicated.
As shown in Fig. 2A, GST-Flag alone was not exported from injected nuclei. Addition of NES1APC to GST-Flag, however, resulted in efficient nuclear export. The NES2APC also directed nuclear export of GST-Flag, with a significant proportion (15%) of cells showing a fairly complete shift of GST-NES2APC to the cytoplasm, and many of the remaining cells showing an increase in cytoplasmic labeling relative to the BSA injection control (Fig. 2A, middle panels). These results indicate that NES1APC and NES2APC are each sufficient to target GST protein for nuclear export. We have also tested NES1APC from rodent APC for its ability to target GST for nuclear export. Although the rodent NES1 sequence does not strictly conform to the NES consensus (Fig. 1B), it did target GST-Flag for nuclear export in 100% of the 237 cells injected (data not shown).
To further examine the functionality of NES1APC and NES2APC, we tested whether either could substitute for the NES sequence in the HIV Rev protein. Chimeric proteins, Rev/NES1 and Rev/NES2, retain the HIV-1 Rev RNA binding and multimerization domains but carry either NES1APC or NES2APC in place of the Rev NES. The constructs were tested for Rev activity by transient transfection of human 293T cells together with a reporter construct, pDM128/CMV (16). pDM128/CMV encodes the cat indicator gene and the HIV-1 Rev response element located between intact 5′ and 3′ splice sites. Nuclear export, and hence expression, of the unspliced CAT mRNA thus requires coexpression of functional Rev protein. As seen in Fig. 2B, this construct induced only a low level of CAT activity when transfected alone into human 293T cells (Neg). However, cotransfection with a plasmid encoding wild-type HIV-1 Rev (Rev), but not with a plasmid encoding an NES-mutant Rev (Rev/M32), greatly enhanced the CAT activity. The chimeric plasmids, Rev/NES1APC and Rev/NES2APC, each significantly enhanced CAT expression. Although the activity induced by the Rev/NES1APC chimera was equivalent to that of wild-type Rev, the Rev/NES2APC was only moderately active. Finally, two Rev/NESAPC chimeras carrying mutated NESAPC (alanines substituted for two critical leucine residues underlined in Fig. 1A, Rev/NES1M and Rev/NES2M) failed to induce CAT activity. Western analysis indicated equivalent expression levels for each of these four chimeric proteins in transfected 293T cells (data not shown).
APC NESs Are Functional Within the Context of the APC Sequence.
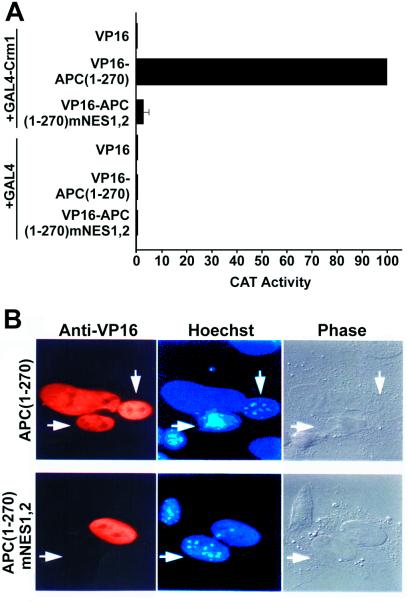
Using a mammalian two-hybrid assay (16), we determined that NES1APC and NES2APC were each able to interact with Crm1 in vivo (data not shown). Crm1 is an NES receptor that functions in the nuclear export of proteins carrying leucine-rich NES sequences. To assess whether NES1APC and NES2APC interact with Crm1 in the context of native APC protein sequence, we again used a mammalian cell two-hybrid system. In this system, CAT protein is expressed from an indicator plasmid only if the APC(1–270) peptide, fused to both a transcription activation domain and an NLS, recruits Crm1, which is fused with a DNA binding domain. Amino acids 1–270 of the APC protein were fused with the VP16 transcription activation domain and the SV40 T antigen nuclear localization signal (VP16-NLSSV40-APC1–270). For analysis, 293T cells were cotransfected with the VP16-NLSSV40-APC1–270 construct, a CAT-based indicator plasmid, pG6(−31)HIVLTRΔTAR, which contains reiterated GAL4 DNA binding sites 5′ to a minimal promoter element (18), and an expression plasmid that encodes a fusion protein consisting of the GAL4 DNA binding domain fused to Crm1 (GAL4/Crm1) (16). This resulted in a ≥200-fold activation in CAT protein expression. A similar transfection but with a VP16 construct instead of the VP16-NLSSV40-APC1–270 construct showed little or no CAT expression (Fig. 3A). Cotransfection with the VP16-NLSSV40-APC1–270 construct containing mutant NESAPC sequences also did not lead to enhanced CAT activity. Western blot analysis demonstrated that the expression levels of the different chimeric proteins were comparable (data not shown). The activation mediated by the VP16-NLSSV40-APC1–270 construct thus reflects recruitment of the VP16 activation domain to the pG6(−31)HIVLTRΔTAR indicator plasmid due to a Crm1–NESAPC interaction, confirming that APC(1–270) interacts with the Crm1 nuclear export factor and that this interaction requires intact NESAPC sequences.
Figure 3.
APC(1–270) interacts with Crm1 and is exported from nuclei. (A) APC(1–270) containing wild-type NES1APC and NES2APC, but not mutant forms, was able to interact with the human Crm1 nuclear export receptor in a mammalian two-hybrid assay. See Results for assay description. Results represent the average of three independent experiments with standard deviation as indicated. (B) Nucleocytoplasmic shuttling of an APC protein domain. A fusion protein consisting of APC residues 1–270 with wild-type [APC(1–270)] or mutant (mNES1,2) forms of NES1APC and NES2APC linked to the VP16 transcription activation domain and the SV40 T antigen NLS was expressed in HeLa cells by transfection. Following fusion to mouse 3T3 cells in the presence of cycloheximide, the resultant heterokaryons were incubated for 2 h, fixed, and then examined by immunofluorescence for nucleocytoplasmic shuttling of the APC fusion protein between the human and murine nuclei. The murine nuclei are identified by their punctate staining with the dye Hoechst 33258 and are indicated by arrows.
To address further whether NES1APC and NES2APC function within the context of native APC protein sequence, the VP16-NLSSV40-APC1–270 construct was analyzed in a heterokaryon assay. VP16-NLSSV40-APC1–270 localizes to the nucleus when expressed in human HeLa cells. HeLa cells transfected with the VP16-NLSSV40-APC1–270 expression plasmid were allowed to synthesize protein for 36 h. These cells were treated with a protein synthesis inhibitor and fused with murine 3T3 cells using polyethylene glycol. If the APC NESs were functional in nuclear export, this chimeric protein would be expected to move from the HeLa nuclei into the cytoplasm and subsequently enter the murine nuclei. As shown in Fig. 3B, by 2 h postfusion the chimeric protein had migrated into the murine nuclei (APC(1–270), arrows). In contrast, a chimeric protein in which the two candidate APC NESs had been mutated (APC(1–270),mNES1,2) failed to migrate from the human nuclei into the murine nuclei. Thus, NES sequences within the context of APC(1–270) direct rapid export from the nucleus.
Cytoplasmic Localization of APC Protein Depends on NES Function.
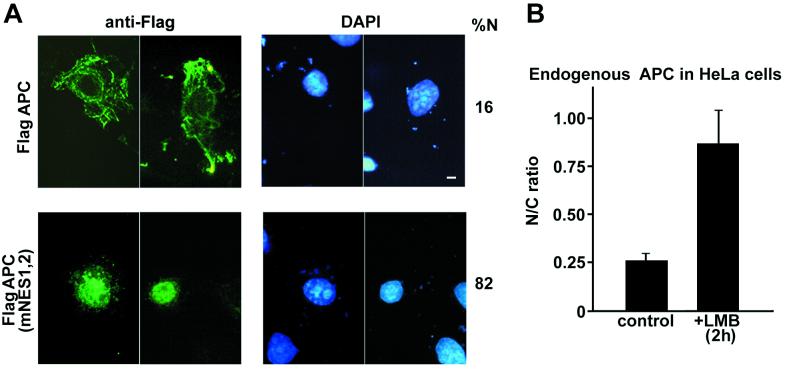
To determine whether the cytoplasmic localization of full-length APC protein depends on the NES1APC and NES2APC sequences, a full-length APC expression construct with an N-terminal Flag epitope tag was introduced into Cos-7 cells by transient transfection for immunofluorescence analysis. As expected, in the great majority (84%) of the transfected cells, Flag-APC protein was seen predominantly in the cytoplasm, often displaying a filamentous staining pattern (Fig. 4A, Flag-APC) as previously described (22, 23). In the remaining 16% of the cells, APC was observed in both cytoplasm and nucleus. If full-length APC shuttles between the nucleus and the cytoplasm, and if the shuttling depends on the NESAPC sequences, mutation of the nuclear export signal(s) should lead to accumulation of nuclear APC. As shown in Fig. 4A, APC protein carrying mutant NES1APC and NES2APC increased the incidence of nuclear staining to 82%. Furthermore, 68% of the cells displayed only nuclear staining, with no visible cytoplasmic staining. Intranuclear staining was confirmed by confocal microscopy (data not shown). To confirm that the various APC proteins had similar expression levels and stabilities, 293T cells were transfected with the various flag-APC constructs, and protein levels were measured in cell lysates. Protein expression levels varied by less than 20% (data not shown). These results demonstrate that NES1APC and NES2APC are necessary for the cytoplasmic localization of full-length APC protein and strongly support their functionality as nuclear export signals.
Figure 4.
Mutation of the NESs in full-length APC results in accumulation of protein in the nucleus. (A) Cos7 cells were transfected with plasmids that express Flag-tagged APC protein (APC) or Flag-APC with mutations in both NES1APC and NES2APC, APC(mNES1,2). At 24 h posttransfection, cells were prepared for immunofluorescence microscopy using an antibody raised against the Flag epitope (green, left panels). Nuclei were visualized using 4′,6-diamidino-2-phenlindole (DAPI) stain (blue, right panel). Scoring 100 cells for FLAG-tagged APC protein distribution revealed that mutation of both NES1APC and NES2APC resulted in accumulation of APC protein in the nucleus. (Scale bar, 10 μm.) (B) LMB treatment leads to an increase in endogenous APC protein in the nuclear fraction of HeLa cells. Following a 2-h treatment with LMB, HeLa cells were fractionated, and proteins from cytosolic and nuclear fractions were separated by SDS/PAGE. APC was identified by Western immunoblot using an ECL substrate. Bands from images captured with a Lumi-imager were quantitated, and values for five independent experiments were expressed as nuclear/cytoplasmic ratio (N/C) of full-length APC protein ±SD.
Endogenous APC Shuttles Between the Nucleus and the Cytoplasm.
To determine whether endogenous APC is exported from the nucleus, the distribution of APC in HeLa cells was examined after treatment with leptomycin B (LMB), a specific inhibitor of Crm1-dependent nuclear export (24). If endogenous APC shuttles between nucleus and cytoplasm, and its nuclear export is dependent on Crm1, then LMB should induce accumulation of APC in the nucleus. HeLa cells treated with LMB were separated into cytosolic and nuclear fractions, and APC protein was quantitated with Western immunoblots. Five independent experiments showed a >3-fold increase in the nuclear/cytoplasmic ratio of full-length APC protein following 8 h of LMB treatment (Fig. 4B). In untreated cells, APC localized to both the cytoplasmic and the nuclear compartments, with a nuclear/cytoplasmic ratio of 0.26. After 8 h of LMB treatment, the nuclear/cytoplasmic ratio was 0.87 (P = 0.0017). This demonstrates that blocking nuclear export by LMB leads to the accumulation of endogenous APC in the nucleus and indicates that endogenous APC shuttles between the nucleus and cytoplasm.
Discussion
We have demonstrated that APC protein shuttles between the nucleus and cytoplasm, a novel activity of potential importance in tumor suppression. The nuclear export of APC protein depends on a functional interaction between the nuclear export factor Crm1 and two amino-terminal nuclear export signals (NES1APC and NES2APC,). Either NESAPC can mediate nuclear export of target RNAs when substituted for the essential Rev NES (Fig. 2B). Each NESAPC is able to interact with Crm1 (data not shown) and mediate nuclear export of a linked GST protein (Fig. 2A). Moreover, these sequences functioned as NESs when examined within their native context. Specifically, the N-terminal 270 aa of APC, which contains both NES1APC and NES2APC, also interacted with Crm1 (Fig. 3A), and this domain was sufficient to confer nuclear export when linked to a heterologous NLS and analyzed using a heterokaryon shuttling assay (Fig. 3B).
Two lines of evidence demonstrate that the wild-type NES sequences are also able to export full-length APC protein. First, APC protein lacking NES1APC and NES2APC displayed a markedly higher level of nuclear localization than wild-type APC protein when expressed in Cos7 cells (Fig. 4A). Second, treatment of HeLa cells with LMB, an inhibitor of leucine-rich NES-dependent nuclear export, led to an increase in the nuclear accumulation of endogenous APC (Fig. 4B). APC localization is thus dynamic, with its predominant steady-state localization in the cytoplasm dependent on the activity of NES1APC and NES2APC.
Subcellular redistribution and compartmental sequestration of proteins have emerged as important mechanisms in the regulation of cellular response. The idea that APC is located in both nucleus and cytoplasm led to speculation that APC might shuttle between these two subcellular compartments (25). Indeed, the present study has demonstrated that APC protein contains two functional, leucine-rich, Crm-1-dependent NESs, which facilitate shuttling of APC protein between the cytoplasm and nucleus. This demonstration leads us to speculate that the nuclear APC protein might impact the Wnt signaling pathway by directly effecting the abundance or activity of nuclear β-catenin, as has been previously shown for cytoplasmic APC and β-catenin.
Acknowledgments
We thank C. Anderson for construction of pGST-APC270 and confocal microscopy assistance, M. Yoshida for leptomycin B, J. Logan for the Flag-APC expression construct, E. Meenan for oligonucleotide synthesis, and K. Ullman, K. Spancake, R. Dawson, S. Prescott, and F. Fitzpatrick for helpful comments on the manuscript. This work was supported by Grants 5PO1 CA73992-02 and DAMD17-96-1-6173 and the Huntsman Cancer Institute (to R.L.W., K.L.N., and F.Z.), by the Howard Hughes Medical Institute (to H.B., Y.K., and B.C.), and by National Institutes of Health Grant GM50877 (to D.A.N. and M.C.B).
Abbreviations
- APC
adenomatous polyposis coli
- NES
nuclear export signal
- LMB
leptomycin B
- NLS
nuclear localization signal
- CMV
cytomegalovirus
- CAT
chloramphenicol acetyltransferase
- GST
glutathione S-transferase
- SV40
simian virus 40
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220401797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220401797
References
- 1.Powell S M, Zilz N, Beazer-Barclay Y, Bryan T M, Hamilton S R, Thibodeau S N, Vogelstein B, Kinzler K W. Nature (London) 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 2.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 4.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 5.Dale T C. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dierick H, Bejsovec A. Curr Top Dev Biol. 1999;43:153–190. doi: 10.1016/s0070-2153(08)60381-6. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi Y, Iwao K, Nawa G, Yoshikawa H, Ochi T, Nakamura Y. Oncol Res. 1998;10:591–594. [PubMed] [Google Scholar]
- 8.Sparks A B, Morin P J, Vogelstein B, Kinzler K W. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 9.Neufeld K L, White R L. Proc Natl Acad Sci USA. 1997;94:3034–3039. doi: 10.1073/pnas.94.7.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klymkowsky M W, Williams B O, Barish G D, Varmus H E, Vourgourakis Y E. Mol Biol Cell. 1999;10:3151–3169. doi: 10.1091/mbc.10.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong M H, Hermiston M L, Syder A J, Gordon J I. Proc Natl Acad Sci USA. 1996;93:9588–9593. doi: 10.1073/pnas.93.18.9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Waltzer L, Bienz M. Nat Cell Biol. 1999;1:144–151. doi: 10.1038/11064. [DOI] [PubMed] [Google Scholar]
- 13.Nathke I S, Adams C L, Polakis P, Sellin J H, Nelson J. J Cell Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nix D A, Beckerle M C. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ailenberg M, Silverman M. BioTechniques. 1997;22:624–630. doi: 10.2144/97224bm11. [DOI] [PubMed] [Google Scholar]
- 16.Bogerd H P, Echarri A, Ross T M, Cullen B R. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malim M H, McCarn D F, Tiley L S, Cullen B R. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Southgate C D, Green M R. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 19.Fridell R A, Bogerd H P, Cullen B R. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hope T J, Huang X J, McDonald D, Parslow T G. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael W M, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 22.Munemitsu S, Souza B, Muller O, Albert I, Rubinfeld B, Polakis P. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- 23.Smith K, Levy D, Maupin P, Pollard T, Vogelstein B, Kinzler K. Cancer Res. 1994;54:3672–3675. [PubMed] [Google Scholar]
- 24.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 25.Bienz M. Curr Opin Genet Dev. 1999;9:595–603. doi: 10.1016/s0959-437x(99)00016-7. [DOI] [PubMed] [Google Scholar]