Abstract
The insulin-like growth factor (IGF) type 1 receptor is required for growth, transformation, and protection from apoptosis. IGFs can enhance cell migration, which is known to be influenced via regulation of the E-cadherin/β-catenin complex. We sought to investigate whether IGF-1 modulated the interaction between E-cadherin and β-catenin in human colorectal cancer cells. We used the C10 cell line, which we established and have previously shown to lack adenomatous polyposis coli, E-cadherin, or β-catenin mutations. We found that IGF-1 stimulation enhanced tyrosine phosphorylation of two proteins, β-catenin and insulin-receptor substrate 1, which formed a complex with E-cadherin. Tyrosine phosphorylation of β-catenin was accompanied by rapid (<1 min) dissociation from E-cadherin at the plasma membrane, followed by relocation to the cellular cytoplasm. IGF-1 also enhanced the stability of β-catenin protein. Despite this, we observed no enhancement of transcriptional activity in complex with T-cell factor 4 (Tcf-4) in human embryonic kidney 293 cells treated with IGF-1 or insulin alone. IGF-1 did, however, enhance transcriptional activity in combination with lithium chloride, an inhibitor of glycogen synthase kinase 3β, which also stabilizes β-catenin. In conclusion, we have shown that IGF-1 causes tyrosine phosphorylation and stabilization of β-catenin. These effects may contribute to transformation, cell migration, and a propensity for metastasis in vivo.
The insulin-like growth factor receptor (IGF1R) is a tetrameric glycoprotein composed of two α and two β subunits. It belongs to the tyrosine kinase receptor superfamily (1). Upon engagement of ligand, the receptor undergoes β-subunit autophosphorylation on tyrosine residues 1131, 1135, and 1136 (2). This results in the recruitment and subsequent tyrosine phosphorylation of intracellular substrates including IRS-1, IRS-2, Shc, and Grb10 (reviewed in ref. 3).
Targeted disruption of the IGF1R in mouse embryos has clarified the role of the IGF1R in normal cell growth and transformation. Mice homozygous for a disrupted IGF1R gene show intrauterine growth retardation and die immediately after birth (4). Cells derived from these embryos, termed R-, are able to grow in media supplemented with 10% serum but do so more slowly than wild-type cells, and each stage of the cell cycle is lengthened. Moreover, R-cells are refractory to transformation by oncogenes including simian virus 40 large T and Ha-ras (5, 6). IGF1R signaling also protects from apoptosis induced by c-myc, by IL-3 withdrawal in hemopoietic cells, and by loss of matrix adhesion in fibroblasts (anoikis; refs. 7–9). In addition, use of a dominant negative mutant receptor in tumors to inhibit IGF1R signaling has been shown to cause massive apoptosis (10).
Various tumor types have been shown to overexpress functional IGF1R including ovarian carcinoma (11) and rhabdomyosarcoma (12). There are conflicting reports, however, regarding levels of IGF1R in colorectal cancer compared with normal mucosa. Zenilman and Graham provided evidence that IGF1R mRNA levels remain unchanged during the transition of colonic epithelium from a normal to a neoplastic state (13). Other reports have suggested overexpression of IGF1R in adenocarcinoma of the colon (14) and an increased number of IGF1Rs during the progression of colorectal adenoma to carcinoma (15).
β-Catenin is a multifunctional molecule that associates with a wide variety of protein partners, including the protein product of the adenomatous polyposis coli (APC) tumor suppressor gene. This interaction regulates free cytoplasmic β-catenin (16, 17). β-Catenin has also been shown to interact with tyrosine kinase receptors (18, 19) at the cell periphery and with transcription factors including the Lef/Tcf family in the nucleus, activating expression of target genes including c-myc (20).
β-Catenin protein can be stabilized by mutational inactivation of the APC gene, usually leading to a truncated protein product, or β-catenin mutations at regulatory amino-terminal serine residues. Both situations lead to nuclear translocation of β-catenin and increased transcriptional activation. Such mutations have been observed in many cancers but have been particularly well documented in colorectal cancer (21–23). Signaling by the growth factor Wnt also leads to cytoplasmic accumulation of β-catenin (24). This occurs via inhibition of signaling by the serine/threonine kinase glycogen synthase kinase 3β (GSK3β), which is a component of the APC/axin/β-catenin complex. GSK3β constitutively phosphorylates amino-terminal residues of β-catenin in the presence of axin (25), resulting in the ubiquitin-mediated proteasomal degradation of β-catenin (26, 27).
β-Catenin is an essential component of adhesion complexes in cells. In epithelial cells, β-catenin binds to the intracellular domain of cadherins, predominantly E-cadherin, linking it to the actin cytoskeleton via α-catenin (28). This cytoskeletal link can be severed by tyrosine phosphorylation of E-cadherin-associated catenins induced by both receptor and nonreceptor tyrosine kinases including the EGF receptor and c-Src (29). This contributes to loss of E-cadherin function resulting in reduced cell–cell adhesion (30). Loss of E-cadherin expression is a common observation in the transition from normal cells to highly malignant human epithelial cancers (31). This may be caused by mutation of the E-cadherin gene itself (32), repression of E-cadherin promoter activity by transcription factors (33, 34), or an increase in promoter methylation (35, 36).
IGF signaling is known to influence integrin-mediated cell motility and adhesion to the substratum (37). We noted that cultured cancer cells expressing antisense IGF1R RNA were less motile and had altered cell–cell contacts (M.P.P. and V.M.M., unpublished observations). Therefore, we investigated whether IGF-1 signaling can influence the function of adherens junctions. The parallels between Wnt signaling and the previously documented inhibition of GSK3 by IGF-1 (38) prompted us to investigate whether signaling by IGF-1 is also a factor in the regulation of β-catenin stability and transcriptional activity.
Materials and Methods
Cell Culture.
All cell lines were maintained in DMEM or RPMI 1640 with 2% bicarbonate with 10% FCS, 100,000 units/liter penicillin, and 100 mg/liter streptomycin sulfate at 37°C in 5% CO2. Cultures were negative for mycoplasma infection.
Ribonuclease Protection Assays.
Total RNA was made using the RNeasy (Qiagen) kit. In all assays, 25 μg of total RNA was hybridized overnight at 60°C using the first 286 bp of the IGF1R cDNA as an antisense riboprobe. Assays were performed as described previously (39).
Immunoprecipitation and Western Blot Analysis.
Subconfluent cultures were serum starved for 24 h, stimulated with serum-free medium containing 1.3 nM (10 ng/ml) IGF-1 (GIBCO/BRL) or water control for the indicated times at 37°C. Cultures were washed in cold PBSA and lysed with Nonidet P-40 lysis buffer (1% Nonidet P-40/150 mM NaCl/50 mM Tris, pH 8.0) or Triton lysis buffer (1% Triton/10 mM Na2HPO4-NaH2PO4, pH 7/150 mM NaCl/5 mM EDTA) including protease inhibitors (10 μg/ml each of pepstatin, aprotinin, leupeptin, and 100 μg/ml phenylmethylsulfonyl fluoride) plus 1:100 phosphatase inhibitor mixture I and II (Sigma). Equal amounts of protein were immunoprecipitated with either an antibody to E-cadherin (clone HECD-1-ICRF) or β-catenin (Transduction Laboratories, Lexington, KY) overnight at 4°C. Following isolation with protein G beads, the immunocomplexes were separated by 7.5% SDS/PAGE, and the proteins were transferred to Hybond ECL (Amersham Pharmacia). The filters were blocked in PBSA with 5% nonfat milk and 0.05% Tween 20 for 1 h and then incubated with primary antibody to β-catenin, IRS-1 (c-20, Santa Cruz Biotechnology), PY99 (Santa Cruz Biotechnology), or E-cadherin for 1–3 h at room temperature. Detection was with secondary antibody conjugated to horseradish peroxidase (Dako) and ECL Plus (Amersham Pharmacia).
Cell Fractionation Experiments.
C10 cells were cultured to 50% confluence, serum starved for 24 h, and then stimulated with 1.3 nM IGF-1 or water control for the indicated times. The cells were disaggregated using PBSA-3 mM EDTA and pellets lysed in digitonin lysis buffer (1% digitonin/150 mM NaCl/50 mM Tris⋅Cl, pH 7.5/10 mM MgCl2) plus protease inhibitors. The lysates were centrifuged at 13,000 rpm for 10 min, and supernatants representing cytosolic components were saved. The pellets representing cytoskeletal and nuclear components were lysed in RIPA buffer (150 mM NaCl/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/50 mM Tris, pH 7.5). Equal amounts of proteins were separated on 7.5% SDS/PAGE gels, transferred to nitrocellulose (Amersham Pharmacia), and immunoblotted for β-catenin.
Immunofluorescence.
C10 cells were plated onto polysine-coated slides and incubated at 37°C overnight. Following 24-hour serum starvation, the cells were stimulated with serum-free medium containing 1.3 nM IGF-1 or vehicle control for 30 min at 37°C. The slides were washed three times in PBSA, and localization of β-catenin was performed as described (40).
Pulse–Chase Analysis.
C10 cells were cultured to 70% confluence. Some cultures were preincubated with 50 mM LiCl for 24 h. The cells were disaggregated using 3 mM EDTA in PBSA. For each chase time point, 2 × 106 cells were washed once in RPMI-1640 without cysteine/methionine (starve medium, Sigma) and resuspended in starve medium and incubated for 1 h at 37°C. Cells were pulsed for 30 min at 37°C using 35S-Promix (Amersham Pharmacia), 10 μCi per 106 cells. The cells were washed three times in starve media in the presence or absence of IGF-1 (6.5 nM), cycloheximide (10 μg/ml) to prevent new protein translation, 15 μg/ml L-methionine and 50 μg/ml L-cysteine (GIBCO/BRL) and chased for the indicated time points. The cells were lysed in RIPA buffer containing protease inhibitors as previously described and immunoprecipitated with β-catenin antibody for 3 h at 4°C. Immunocomplexes were isolated with protein G and separated on 7.5% SDS/PAGE gels, stained with Coomassie blue (Sigma), destained and incubated with Amplify (Amersham Pharmacia) fluorographic reagent, dried, and exposed to film.
Luciferase Reporter Assays.
HEK293 cells (1.5 × 107) were plated in 15-cm dishes and incubated overnight at 37°C. The cells were transfected with 13.7 μg each of TOPFLASH or FOPFLASH reporter plasmids (21), human-Tcf-4, and β-catenin expression vectors using 18.3 μl of Lipofectamine (GIBCO/BRL). On the following day, the cells were replated at 5 × 105 into 6-well plates and incubated at 37°C for 8 h. The cells were washed in serum-free medium and serum starved overnight in the presence or absence of LiCl at the indicated concentration for 16 h. Cultures were incubated with IGF-1 or insulin at 6.5 nM (50 ng/ml) for the indicated times. Reporter assays were performed using the luciferase reporter system (Promega) on a TD 20/20 luminometer (Turner Designs).
Results
Analysis of IGF1R in Colorectal Cancer Cell Lines.
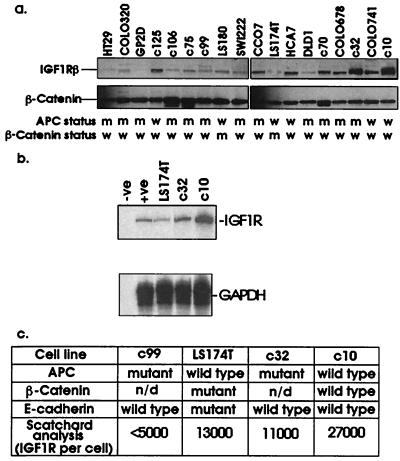
A panel of 20 colorectal cancer cell lines was assessed for their level of expression of IGF1R protein and mRNA levels. Many of these cell lines were low-passage cell lines derived from biopsies of invasive colorectal cancers. These cell lines have been described in detail elsewhere (41). We observed a considerable variation in IGF1R expression between colorectal cell lines. IGF1R expression at the RNA level correlated positively with that of IGF1R protein (Fig. 1 a and b). We noted that four cell lines, which lacked APC and β-catenin mutations, tended to have the highest receptor levels. The C10 cell line had higher levels of IGF1R than the other cell lines tested and had approximately 27,000 sites per cell using Scatchard analysis. This cell line also lacks E-cadherin and p53 mutations. C10 IGF1R were deemed functional due to their enhanced proliferation in response to exogenous IGF-1 (data not shown). Hence, we used this cell line to study IGF-1 effects on β-catenin localization and stability.
Figure 1.
Analysis of IGF1R levels in human colorectal cancer cell lines. Lysates (20 μg) were analyzed for IGF1R and β-catenin levels by Western blotting using an antibody against the β-subunit of the receptor, and filters were reprobed for β-catenin (a). Levels of IGF1R RNA were analyzed by ribonuclease protection assay using the first 286 bases of the IGF1R as an antisense riboprobe (b). Subsequently, detailed analysis of the levels of IGF1R per cell was carried out by Scatchard analysis using 125I-IGF-1 (c). Information on the mutational status of APC, β-catenin, and E-cadherin was taken from ref. 41. n/d, Not done.
The Effects of IGF-1 Signaling on the Interaction Between β-Catenin and E-Cadherin.
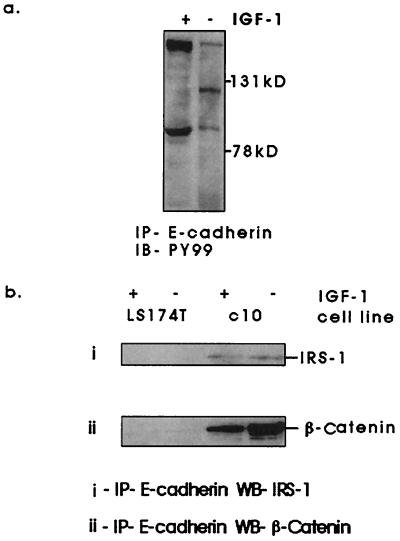
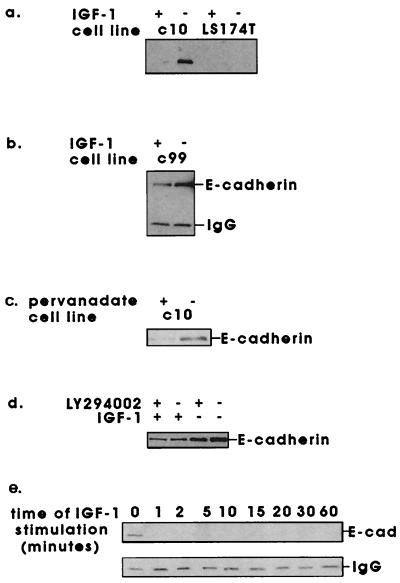
β-Catenin has been reported to interact directly with the receptor tyrosine kinase epidermal growth factor and c-erbB-2 (18, 19). We found no evidence of interaction with the IGF1R as assessed by coimmunoprecipitation between β-catenin and IGF1R or its principal substrate IRS-1 (data not shown). IGF-1 stimulation of C10 cells did, however, lead to a differential pattern of tyrosine phosphorylation of proteins that coimmunoprecipitated with E-cadherin (Fig. 2a). Two proteins of approximate molecular mass of 90 and 180 kDa responded to IGF-1 stimulation with enhanced tyrosine phosphorylation. Proteins of equivalent molecular mass were subsequently identified in E-cadherin immunoprecipitates as β-catenin and IRS-1 (Fig. 2b). We also noted a 120-kDa protein, which responded to IGF-1 stimulation with reduced tyrosine phosphorylation. This was likely to be either E-cadherin itself or another member of the catenin family, p120ctn. Coimmunoprecipitation between E-cadherin and p120ctn has been observed in this cell line (data not shown). The interaction between IRS-1 and E-cadherin seemed to be independent of IGF-1 stimulation. This was not the case for β-catenin and E-cadherin (Fig. 2 a and b). Fig. 3a shows a converse immunoprecipitation and Western blot following IGF-1 stimulation. The interaction between β-catenin and E-cadherin was maintained in serum-free conditions but was lost upon stimulation with IGF-1. We could detect no interaction between E-cadherin and β-catenin in LS174T cells, which have two E-cadherin gene mutations and may express truncated E-cadherin protein lacking β-catenin binding sites (32). We saw similar effects in other cell lines with lower levels of functional IGF1R (Fig. 3b), including C99 (41), which carries an APC mutation and exhibits approximately 5,000 IGF1R per cell (Fig. 1c).
Figure 2.
Effects of IGF-1 stimulation on tyrosine phosphorylation of proteins that coimmunoprecipitate with E-cadherin. Cells were serum starved for 16 h and treated with recombinant 1.3 nM (10 ng/ml) IGF-1 for 10 min. Cells were lysed in Nonidet P-40 lysis buffer, and proteins were immunoprecipitated using anti-E-cadherin antibody (clone HECD-1). Immunoprecipitates were assessed for tyrosine phosphorylation (a) using phosphotyrosine antibody (PY99). Two proteins of approximate molecular mass of 180 kDa and 90 kDa were identified by Western blotting as insulin receptor substrate-1 (IRS-1) and β-catenin (b).
Figure 3.
Effects of IGF-1 stimulation on the interaction between β-catenin and E-cadherin. Cells were serum starved for 16 h, then stimulated with 1.3 nM (10 ng/ml) IGF-1 for 10 min (a and b), pervanadate (0.5 mM sodium orthovanadate and 1.5 mM hydrogen peroxide) for 30 min (c), 1.3 nM IGF-1 for 10 min following preincubation with 100 μM LY294002 or DMSO carrier for 30 min (d), or with 1.3 nM IGF-1 from 1 to 60 min (e), all at 37°C. Cells were lysed in Triton lysis buffer, and the interaction between β-catenin and E-cadherin was examined by immunoprecipitation of β-catenin and Western blotting for E-cadherin.
These findings appeared to link enhanced tyrosine phosphorylation with disruption of adherens junctions. We anticipated that the effects could be mimicked by inhibition of tyrosine phosphatases. Fig. 3c shows that treatment with pervanadate also induced disruption of the β-catenin/E-cadherin complex in C10 cells. This effect was not seen with the serine phosphatase inhibitor okadaic acid, which, used at 1 μM, inhibits protein phosphatases 1 and 2A (data not shown). Similarly, this effect of IGF-1 was not blocked by inhibitors of phosphatidylinositol 3-kinase (LY294002, Fig. 3d). The dissociation of the E-cadherin/β-catenin complex was rapid, occurring in under 1 min at 37°C, and was sustained for over 1 h (Fig. 3e).
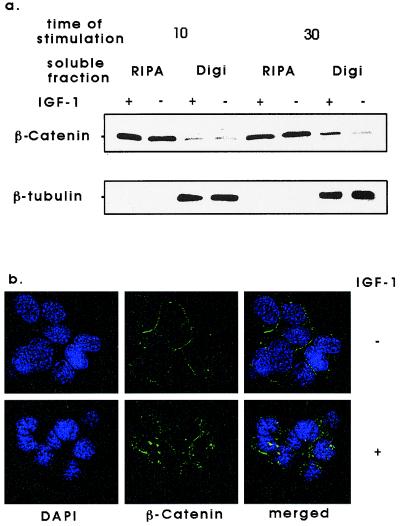
We next investigated the effects of IGF-1 stimulation on the localization of β-catenin in C10 cells. After 10- and 30-min IGF-1 stimulation, cell pellets were lysed in digitonin lysis buffer to solubilize cytoplasmic proteins. Digitonin-insoluble nuclear and cytoskeletal proteins were solubilized in RIPA buffer. Immunoblot analysis for β-catenin showed that after 10 min of IGF-1 treatment, there was no apparent difference in the cellular localization of β-catenin. After 30 min of IGF-1 treatment, β-catenin appeared to partially relocate from RIPA to digitonin-soluble components (Fig. 4a). Consistent with this, immunofluorescence staining of C10 cells for β-catenin after 30 min of IGF-1 treatment showed that β-catenin moved from a peripheral location to a punctate cytoplasmic distribution (Fig. 4b). These results confirm that IGF-1 stimulation severed the link between E-cadherin and the actin cytoskeleton, with the relocation of β-catenin from a submembrane to a cytoplasmic location.
Figure 4.
Effects of IGF-1 stimulation on the localization of β-catenin in c10 cells. (a) Subconfluent c10 cells were serum starved for 16 h and were treated with 1.3 nM IGF-1 for 10 or 30 min. Cells were fractionated into 1% digitonin or RIPA-soluble fractions, and equal amounts of each soluble fraction were separated by SDS/PAGE and immunoblotted for β-catenin. (b) Subconfluent c10 cells were serum starved for 16 h and were treated with 1.3 nM IGF-1 or vehicle control for 30 min at 37°C. Cells were stained with an antibody for β-catenin that was visualized with a fluorescent secondary antibody. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). (×400.)
The Effects of IGF-1 Signaling on Stability and Transactivation Potential of β-Catenin.
Next, we investigated the functional consequences of the IGF-1-induced change in β-catenin localization. IGF-1 is known to inhibit GSK3β (42). Because accumulation of β-catenin in a cadherin-free pool correlates with TCF-induced activation of transcription (43), we postulated that IGF-1 effects on GSK3β could stabilize β-catenin. We confirmed that IGF-1 stimulation could phosphorylate GSK3β on serine 9 and hence inactivate it by performing in vitro kinase reactions in C10 cells. Upon IGF-1 stimulation, immunoprecipitated p90rsk2 induced 2–3-fold phosphorylation of a peptide encompassing the first 15 residues of GSK3β. In both C10 and 293 cell lines, IGF-1-induced phosphorylation was abolished when serine 9 was replaced by alanine (data not shown).
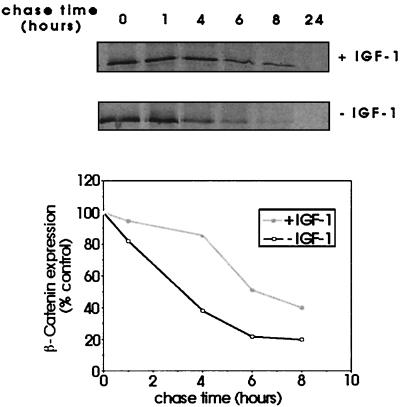
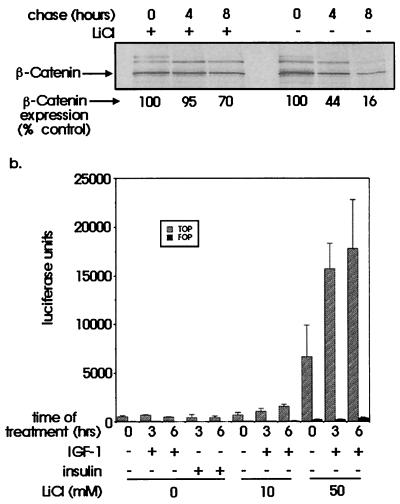
We examined IGF-1 effects on the stability of β-catenin by pulse–chase analysis of C10 cells. When IGF-1 was added at the chase stage, we observed enhancement of β-catenin stability from a half-life of 3 h in serum-free conditions to 6 h with IGF-1 (Fig. 5). As lithium chloride has been shown to inhibit GSK3 noncompetitively, we sought to investigate the effects of this inhibitor on the stability of β-catenin protein. Pulse–chase analysis was performed on C10 cells in the presence or absence of 50 mM LiCl (Fig. 6a). Densitometric analysis indicated that the half-life of β-catenin in cells treated with 50 mM LiCl plus IGF-1 now exceeded 8 h. Coimmunoprecipitating proteins, which migrated more slowly in this figure, were likely to be α-catenin and E-cadherin (Fig. 6a).
Figure 5.
Analysis of effects of IGF-1 signaling on stability of β-catenin protein. Subconfluent C10 cells were pulsed with 35S-Promix and chased with medium containing an excess of cold methionine/cysteine for the indicated times. Cells were lysed and immunoprecipitated for β-catenin. The results were analyzed by densitometry and expressed graphically as a percentage of the value at time 0 h. The figure shows results of a single experiment, which was repeated once with similar results.
Figure 6.
(a) Effects of lithium chloride on stability of β-catenin. Subconfluent C10 cells were serum starved for 16 h in the presence or absence of 50 mM LiCl and then pulsed with 35S-Promix for 30 min and chased with medium containing excess of cold methionine/cysteine. Cells were lysed and immunoprecipitated for β-catenin and analyzed as before. The numbers shown indicate a mean of duplicate experiments. (b). Effects of IGF-1 and lithium chloride on transcriptional activity of β-catenin. HEK293 cells were cotransfected with equal amounts of expression plasmids containing full-length wild-type β-catenin, Tcf-4, and either TOPFLASH or FOPFLASH reporter. Total cell lysates were equalized for protein concentration and assayed for luciferase activity. Bars indicate the mean and standard error of triplicate luciferase assays.
To investigate whether IGF-1 also enhanced transactivation potential, a reporter gene assay was performed in HEK293 cells. We used a reporter construct incorporating multimeric Lef/Tcf promoter sequences upstream of a luciferase reporter (TOPFLASH) and a control construct containing mutated promoter sequences (FOPFLASH, ref. 21). We cotransfected TOPFLASH or FOPFLASH, β-catenin, and human wild-type Tcf-4 wild-type expression constructs into 293 cells. Optimized luciferase assays showed a basal level of TOP/FOP activity of between 4:1 and 20:1. Forty-eight hours after transfection, serum-starved 293 cells were treated with IGF-1 or insulin for 3 or 6 h. These factors alone failed to alter basal reporter activity (Fig. 6b). A 16-h pretreatment of the transfectants with LiCl had a dramatic effect on reporter activity, with significant enhancement at 50 mM LiCl (P < 0.01 by Tukey's test for comparison with activity in the absence of LiCl). We saw a further increase in reporter activity when we added 50 ng/ml (6.5 nM) IGF-1 to transfectants pretreated with LiCl at 10 mM (P < 0.01 after 6 h of IGF-1 treatment) and at 50 mM (P < 0.01 after 3 h of IGF-1 treatment and P < 0.001 after 6 h).
Discussion
β-Catenin interacts with three major cell systems that regulate its fate and function (reviewed in ref. 28). First, β-catenin is a component of adherens junctions where it links cadherins to the actin cytoskeleton. Second, its degradation is regulated by complexing with APC/axin and GSK3β; third, its transcriptional activity is regulated in a nuclear complex with Lef/Tcf transcription factors. We have addressed the role of IGFs in each of these aspects of β-catenin function.
We have shown that treatment of C10 human colorectal cancer cells with IGF-1 enhanced tyrosine phosphorylation of proteins coprecipitating with E-cadherin, namely IRS-1 and β-catenin. Complexes including IRS-1 and E-cadherin have been reported previously in MCF-7 cells (44).
IGF-1-induced tyrosine phosphorylation of β-catenin resulted in rapid disruption of β-catenin from E-cadherin. The speed of this effect (<1 min) argued against involvement of downstream signaling intermediates, such as phosphatidylinositol 3-kinase, given that maximum activity of these kinases after mitogen stimulation occurs after 1 min. Indeed, IGF-1-induced down-regulation of the β-catenin/E-cadherin complex was not blocked with inhibitors of this pathway. Members of the Src family of nonreceptor tyrosine kinases have been shown to phosphorylate β-catenin on tyrosine and disrupt the complex with E-cadherin (45). It is plausible that IGF-1 may recruit and activate c-Src proteins, which are enriched in adherens junctions. However, the demonstration that an inhibitor of c-Src activity, C-terminal Src kinase, can interact directly with the IGF1R argues against this possibility (46). Alternatively, IGF-1 signaling may lead to inactivation of a tyrosine phosphatase. IRS-1, for example, has been shown to directly interact with SHPTP-2 (47), a phosphatase with a role in cell motility (48, 49). The tyrosine phosphatase LAR (leukocyte common antigen-related) has been detected in the E-cadherin/β-catenin complex and has a role in epithelial cell migration (50).
Consistent with our findings, Andre et al. (51) have recently demonstrated enhanced tyrosine phosphorylation of β-catenin upon IGF-1 stimulation in an APC-mutated colorectal cancer cell line, HT29. This resulted in down-regulation of E-cadherin and increased cell motility. This effect of IGFs upon the interaction between β-catenin and E-cadherin may serve to inhibit the function of E-cadherin, which, in vivo could lead to a propensity for metastasis.
We found that a 30-min stimulation of C10 cells with IGF-1 resulted in the relocation of β-catenin from a submembrane to cytoplasmic location. Other growth factors, such as hepatocyte growth factor and intestinal trefoil factor, have been shown to increase the levels of β-catenin-free pools caused by tyrosine phosphorylation, which resulted in concomitant cell migration (52, 53). The secreted factor Wnt-1 also enhanced β-catenin-free pools, not via tyrosine phosphorylation but by suppression of GSK3β leading to increased stability of β-catenin (24, 54). Furthermore, signaling by Wnt enhanced transcriptional activation of β-catenin in complexes with Tcf/Lef transcription factors (55, 56).
We observed an approximate doubling of the half-life of β-catenin in the presence of IGF-1 from 3 to 6 h. However, no β-catenin/Tcf transcriptional activation was observed in the presence of IGF-1 alone. Lithium chloride has been shown to inhibit GSK3β by a mechanism independent of serine 9 phosphorylation (57). We found that LiCl alone at 50 mM increased the half-life of β-catenin protein to a greater extent than that seen with IGF-1 alone, to over 8 h. In addition, LiCl treatment produced a dramatic increase in β-catenin/Tcf transcriptional activation. A combination of LiCl and IGF-1 further increased this transcriptional activity.
In conclusion, LiCl-mediated inhibition of GSK3β appeared to stabilize β-catenin to allow accumulation of free cytoplasmic β-catenin to a sufficient extent to induce Lef/Tcf activation. It appeared that IGF-1 alone did not allow the accumulation of free β-catenin to a threshold sufficient to initiate transcriptional activation. Interestingly, transfection of APC in SW480 cells can still down-regulate β-catenin levels in the presence of 40 mM lithium, suggesting other kinases apart from GSK3β may be involved in the actions of APC (58). As IGF-1 was found to enhance transcriptional activation in the presence of LiCl, it is possible that this unknown kinase might be downstream of signaling from the IGF1R.
Finally, given that APC and GSK3 function as pro-apoptotic signals (21, 59, 60), and excess β-catenin expression protects from apoptosis caused by anoikis (61), we suggest that IGF effects on β-catenin could also explain, at least in part, the anti-apoptotic effects of IGF signaling.
Acknowledgments
We are grateful to Prof. Hans Clevers, Dr. Marc van de Wetering, and Prof. Renato Baserga for providing plasmids, Katja Schmeiser for assistance with confocal microscopy, and Prof. A. L. Harris for encouraging discussions. This study was supported by the Medical Research Council and the Imperial Cancer Research Fund.
Abbreviations
- Tcf-4
T-cell factor 4
- IGF
insulin-like growth factor
- IGF1R
IGF-1 receptor
- APC
adenomatous polyposis coli
- GSK3β
glycogen synthase kinase 3β
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210394297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210394297
References
- 1.van der Geer P, Hunter T, Lindberg R A. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 2.Kato H, Faria T N, Stannard B, Roberts C T, Jr, LeRoith D. Mol Endocrinol. 1994;8:40–50. doi: 10.1210/mend.8.1.7512194. [DOI] [PubMed] [Google Scholar]
- 3.Baserga R, Hongo A, Rubini M, Prisco M, Valentinis B. Biochim Biophys Acta. 1997;1332:F105–F126. doi: 10.1016/s0304-419x(97)00007-3. [DOI] [PubMed] [Google Scholar]
- 4.Liu J P, Baker J, Perkins A S, Robertson E J, Efstratiadis A. Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 5.Sell C, Rubini M, Rubin R, Liu J P, Efstratiadis A, Baserga R. Proc Natl Acad Sci USA. 1993;90:11217–11221. doi: 10.1073/pnas.90.23.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sell C, Dumenil G, Deveaud C, Miura M, Coppola D, Deangelis T, Rubin R, Efstratiadis A, Baserga R. Mol Cell Biol. 1994;14:3604–3612. doi: 10.1128/mcb.14.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrington E A, Bennett M R, Fanidi A, Evan G I. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peruzzi F, Prisco M, Dews M, Salomoni P, Grassilli E, Romano G, Calabretta B, Baserga R. Mol Cell Biol. 1999;19:7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valentinis B, Morrione A, Peruzzi F, Prisco M, Reiss K, Baserga R. Oncogene. 1999;18:1827–1836. doi: 10.1038/sj.onc.1202471. [DOI] [PubMed] [Google Scholar]
- 10.Dambrosio C, Ferber A, Resnicoff M, Baserga R. Cancer Res. 1996;56:4013–4020. [PubMed] [Google Scholar]
- 11.Resnicoff M, Ambrose D, Coppola D, Rubin R. Lab Invest. 1993;69:756–760. [PubMed] [Google Scholar]
- 12.Shapiro D N, Jones B G, Shapiro L H, Dias P, Houghton P J. J Clin Invest. 1994;94:1235–1242. doi: 10.1172/JCI117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenilman M E, Graham W. Cancer Invest. 1997;15:1–7. doi: 10.3109/07357909709018911. [DOI] [PubMed] [Google Scholar]
- 14.Freier S, Weiss O, Eran M, Flyvbjerg A, Dahan R, Nephesh I, Safra T, Shiloni E, Raz I. Gut. 1999;44:704–708. doi: 10.1136/gut.44.5.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakam A, Yeatman T J, Lu L, Mora L, Marcet G, Nicosia S V, Karl R C, Coppola D. Hum Pathol. 1999;30:1128–1133. doi: 10.1016/s0046-8177(99)90027-8. [DOI] [PubMed] [Google Scholar]
- 16.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 17.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoschuetzky H, Aberle H, Kemler R. J Cell Biol. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanai Y, Ochiai A, Shibata T, Oyama T, Ushijima S, Akimoto S, Hirohashi S. Biochem Biophys Res Commun. 1995;208:1067–1072. doi: 10.1006/bbrc.1995.1443. [DOI] [PubMed] [Google Scholar]
- 20.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, Dacosta L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 21.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 22.Ilyas M, Tomlinson I P, Rowan A, Pignatelli M, Bodmer W F. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 24.Papkoff J. J Biol Chem. 1997;272:4536–4543. [PubMed] [Google Scholar]
- 25.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orford K, Crockett C, Jensen J P, Weissman A M, Byers S W. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 27.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benzeev A, Geiger B. Curr Opin Cell Biol. 1998;10:629–639. doi: 10.1016/s0955-0674(98)80039-2. [DOI] [PubMed] [Google Scholar]
- 29.Daniel J M, Reynolds A B. BioEssays. 1997;19:883–891. doi: 10.1002/bies.950191008. [DOI] [PubMed] [Google Scholar]
- 30.Ozawa M, Kemler R. J Biol Chem. 1998;273:6166–6170. doi: 10.1074/jbc.273.11.6166. [DOI] [PubMed] [Google Scholar]
- 31.Christofori G, Semb H. Trends Biochem Sci. 1999;24:73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 32.Efstathiou J A, Liu D, Wheeler J M, Kim H C, Beck N E, Ilyas M, Karayiannakis A J, Mortensen N J, Kmiot W, Playford R J, et al. Proc Natl Acad Sci USA. 1999;96:2316–2321. doi: 10.1073/pnas.96.5.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cano A, Perez-Moreno M A, Rodrigo I, Locascio A, Blanco M J, del Barrio M G, Portillo F, Nieto M A. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 34.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 35.Graff J R, Gabrielson E, Fujii H, Baylin S B, Herman J G. J Biol Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- 36.Wheeler J M D, Kim H-C, Efstathiou J A, Ilyas M, Mortensen N J M, Bodmer W F. Gut. 2000;46:A53. , in press. [Google Scholar]
- 37.Leventhal P S, Feldman E L. Trends Endocrinol Metab. 1997;8:1–6. doi: 10.1016/s1043-2760(96)00202-0. [DOI] [PubMed] [Google Scholar]
- 38.Cross D A, Alessi D R, Vandenheede J R, McDowell H E, Hundal H S, Cohen P. Biochem J. 1994;303:21–26. doi: 10.1042/bj3030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith K, Bui T D, Poulsom R, Kaklamanis L, Williams G, Harris A L. Br J Cancer. 1999;81:496–502. doi: 10.1038/sj.bjc.6690721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmeiser K, Hammond E M, Roberts S, Grand R J A. FEBS Lett. 1998;433:51–57. doi: 10.1016/s0014-5793(98)00850-3. [DOI] [PubMed] [Google Scholar]
- 41.Rowan A J, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer W F, Tomlinson I P. Proc Natl Acad Sci USA. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutherland C, Leighton I A, Cohen P. Biochem J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubinfeld B, Robbins P, Elgamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 44.Guvakova M A, Surmacz E. Exp Cell Res. 1997;231:149–162. doi: 10.1006/excr.1996.3457. [DOI] [PubMed] [Google Scholar]
- 45.Roura S, Miravet S, Piedra J, deHerreros A G, Dunach M. J Biol Chem. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- 46.Arbet-Engels C, Tartare-Deckert S, Eckhart W. J Biol Chem. 1999;274:5422–5428. doi: 10.1074/jbc.274.9.5422. [DOI] [PubMed] [Google Scholar]
- 47.Sun X J, Crimmins D L, Myers M G, Jr, Miralpeix M, White M F. Mol Cell Biol. 1993;13:7418–7428. doi: 10.1128/mcb.13.12.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manes S, Mira E, Gomez-Mouton C, Zhao Z J, Lacalle R A, Martinez A C. Mol Cell Biol. 1999;19:3125–3135. doi: 10.1128/mcb.19.4.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu D H, Qu C K, Henegariu O, Lu X, Feng G S. J Biol Chem. 1998;273:21125–21131. doi: 10.1074/jbc.273.33.21125. [DOI] [PubMed] [Google Scholar]
- 50.Muller T, Choidas A, Reichmann E, Ullrich A. J Biol Chem. 1999;274:10173–10183. doi: 10.1074/jbc.274.15.10173. [DOI] [PubMed] [Google Scholar]
- 51.Andre F, Rigot V, Thimonier J, Montixi C, Parat F, Pommier G, Marvaldi J, Luis J. Int J Cancer. 1999;83:497–505. doi: 10.1002/(sici)1097-0215(19991112)83:4<497::aid-ijc11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 52.Hiscox S, Jiang W G. Anticancer Res. 1999;19:509–517. [PubMed] [Google Scholar]
- 53.Liu D, Elhariry I, Karayiannakis A J, Wilding J, Chinery R, Kmiot W, McCrea P D, Gullick W J, Pignatelli M. Lab Invest. 1997;77:557–563. [PubMed] [Google Scholar]
- 54.Cook D, Fry M J, Hughes K, Sumathipala R, Woodgett J R, Dale T C. EMBO J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]
- 55.Young C S, Kitamura M, Hardy S, Kitajewski J. Mol Cell Biol. 1998;18:2474–2485. doi: 10.1128/mcb.18.5.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu S C, Galceran J, Grosschedl R. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stambolic V, Ruel L, Woodgett J R. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 58.Easwaran V, Song V, Polakis P, Byers S. J Biol Chem. 1999;274:16641–16645. doi: 10.1074/jbc.274.23.16641. [DOI] [PubMed] [Google Scholar]
- 59.Bijur G N, De Sarno P, Jope R S. J Biol Chem. 2000;275:7583–7590. doi: 10.1074/jbc.275.11.7583. [DOI] [PubMed] [Google Scholar]
- 60.Pap M, Cooper G M. J Biol Chem. 1998;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 61.Orford K, Orford C C, Byers S W. J Cell Biol. 1999;146:855–867. doi: 10.1083/jcb.146.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]