Abstract
We introduce a computational method for identifying subcellular locations of proteins from the phylogenetic distribution of the homologs of organellar proteins. This method is based on the observation that proteins localized to a given organelle by experiments tend to share a characteristic phylogenetic distribution of their homologs—a phylogenetic profile. Therefore any other protein can be localized by its phylogenetic profile. Application of this method to mitochondrial proteins reveals that nucleus-encoded proteins previously known to be destined for mitochondria fall into three groups: prokaryote-derived, eukaryote-derived, and organism-specific (i.e., found only in the organism under study). Prokaryote-derived mitochondrial proteins can be identified effectively by their phylogenetic profiles. In the yeast Saccharomyces cerevisiae, 361 nucleus-encoded mitochondrial proteins can be identified at 50% accuracy with 58% coverage. From these values and the proportion of conserved mitochondrial genes, it can be inferred that ≈630 genes, or 10% of the nuclear genome, is devoted to mitochondrial function. In the worm Caenorhabditis elegans, we estimate that there are ≈660 nucleus-encoded mitochondrial genes, or 4% of its genome, with ≈400 of these genes contributed from the prokaryotic mitochondrial ancestor. The large fraction of organism-specific and eukaryote-derived genes suggests that mitochondria perform specialized roles absent from prokaryotic mitochondrial ancestors. We observe measurably distinct phylogenetic profiles among proteins from different subcellular compartments, allowing the general use of prokaryotic genomes in learning features of eukaryotic proteins.
Mitochondria, chloroplasts, and perhaps other cellular organelles have apparently descended from microbes captured by the progenitors of modern eukaryotic cells (1, 2). In time, the genes of these organelles were shifted into the nuclear genome, and transport systems were established to shuttle the nucleus-encoded organellar proteins from the cytoplasm into the organelles (reviewed in ref. 3). Contemporary mitochondrial genomes encode only a few genes, for example, fewer than 20 in yeast (see the Saccharomyces Genome Database, Stanford University, http://genome-www.stanford.edu/Saccharomyces); these genes predominantly encode large integral membrane proteins that are perhaps difficult to transport. The overall number of nucleus-encoded mitochondrial genes is unknown.
Consistent with the prokaryotic heritage inferred from organellar morphology (1, 2), many of the proteins that function in these organelles have molecular properties that more closely resemble prokaryotic proteins than eukaryotic proteins. For example, chloroplast and mitochondrial proteins have average lengths similar to proteins in prokaryotes and are composed of repeating domains with a frequency close to that seen in prokaryotes (4). The amino acid composition of mitochondrial proteins also resembles that of prokaryotic proteins (5), and mitochondrial proteins have been observed to have many homologs among the prokaryotes (6, 7). These observations add to the body of evidence that these organelles are derived from microbes; in the method adopted here, these observations provide a quantitative basis for identifying which proteins encoded by a eukaryotic chromosome are eventually destined for these organelles.
Here, we show that nucleus-encoded proteins destined for different subcellular locations have measurably distinct phylogenetic distributions of homologs. This phylogeny can be described with a phylogenetic profile (8) that specifies the pattern of occurrence of a given protein among organisms with sequenced genomes. Each phylogenetic profile is an ordered list of numbers describing the degree of similarity between the query protein and the best sequence match in each of the sequenced genomes. Previously, it has been shown that proteins with similar phylogenetic profiles often have similar functions (8). Here, we show that the phylogenetic profiles of proteins with the same cellular location are often similar. Hence, this similarity of profiles can be used to assign query proteins to subcellular locations. We focus on mitochondrial proteins.
Methods
Calculating Phylogenetic Profiles.
The analysis was performed for each protein encoded by the open reading frames (ORFs) of the complete genomes of the yeast Saccharomyces cerevisiae [6,217 ORFs (9)] and the worm Caenorhabditis elegans [17,123 ORFs (10)]. Phylogenetic profiles (8) were calculated as in ref. 11 by performing a blast (12) sequence homology search between each yeast or worm protein and the proteins encoded by each of the 31 other fully sequenced genomes (listed in Fig. 1A, available from the National Center for Biotechnology Information Entrez Genomes web site, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = Genome). The value at each position of the phylogenetic profile is equal to −1/log E, where E is the blast expectation value of the best matching protein sequence in a genome, calculated only for alignments with expectation values less than 1 × 10−6 and equal to 1.0 otherwise. In the simplest case, a phylogenetic profile would be a string of zeros for perfect matches and ones for no matches.
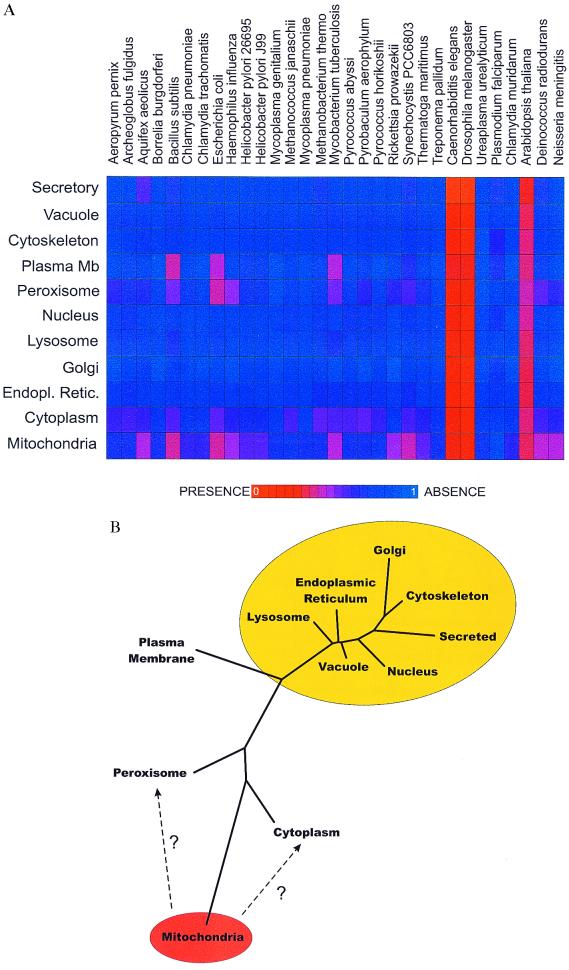
Figure 1.
(A) The mean phylogenetic profile (8) of yeast proteins experimentally localized (14) to different cellular locations. Each profile (a horizontal bar of 31 elements) shows the distribution among genomes of homologs of proteins from one subcellular location. Plasma Mb, plasma membrane. Colors express the average degree of sequence similarity of proteins in that organelle to their sequence homologs in the indicated genomes, with red indicating greater average similarity and blue indicating less, calculated as in the text. Only proteins with at least one homolog among the genomes listed are included. The genomes of Plasmodium and Arabidopsis are only partially complete (≈15% and 50%, respectively). (B) A tree of the observed relationships among the yeast proteins from different subcellular compartments. Overlaid on the tree is our interpretation of the relationships, showing ellipses clustering compartments thought to be derived from the progenitor of mitochondria (orange ellipse) and of the eukaryote nucleus (yellow ellipse). Only proteins with a homolog among the genomes listed in A are examined. A distance matrix was calculated of pairwise Euclidian distances between the mean phylogenetic profiles (A) of proteins known to be localized in each compartment. A tree was generated from this matrix by the neighbor-joining method implemented in PHYLIP 3.5C (J. Felsenstein, University of Washington, Seattle).
Assigning Subcellular Location.
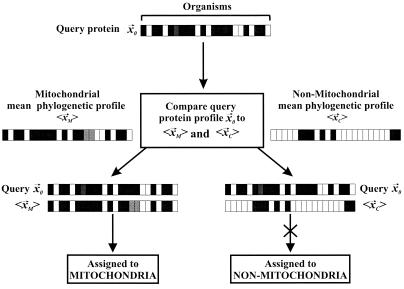
Proteins are assigned to either the mitochondrion or nonmitochondrial cellular location with the use of a linear discrimination function that allows one to decide which distribution, known mitochondrial proteins (XM) or known nonmitochondrial proteins (XC), a given query phylogenetic profile (x⃗0) better matches (Fig. 2). The set XM contains NM phylogenetic profiles (x⃗M,1, x⃗M,2, …, x⃗M,NM) of mitochondrial proteins, and the set XC contains NC phylogenetic profiles (x⃗C,1, x⃗C,2, …, x⃗C,NC) of nonmitochondrial proteins, with each profile derived from n genomes. For the distributions XM and XC, the mean profiles (〈x⃗M〉 and 〈x⃗C〉) and the common covariance matrix S for the two distributions are calculated. The covariance matrix S is an n × n matrix whose elements Si,j, where i and j are indices over the n genomes, are calculated as Si,j = (1/N)∑k=1N(xk,i − 〈xi〉)(xk,j − 〈xj〉), where N = NM + NC and 〈xi〉 = (1/N)∑k=1Nxk,i. The covariance matrix S is calculated by using all N phylogenetic profiles in XM and XC. The linear discrimination function (〈x⃗M〉 − 〈x⃗C〉)T⋅S−1⋅x⃗0, in which T represents the transpose operation, is evaluated to give a numerical result y. The protein with vector x⃗0 is assigned to distribution XM if y exceeds a threshold value t, the midway distance between the two distributions, where t is calculated as ½(〈x⃗M〉 − 〈x⃗C〉)T⋅S−1⋅(〈x⃗M〉 + 〈x⃗C〉); if y < t, x⃗0 is classified into distribution XC. Varying the threshold by a threshold offset (Δt) allows us to increase prediction accuracy (fraction of correct assignments to the mitochondrial category) at the expense of coverage (fraction of known mitochondrial proteins correctly assigned), and vice versa.
Figure 2.
The scheme by which proteins are classified into mitochondrial or nonmitochondrial cellular localizations. Each horizontal bar is a phylogenetic profile; that for the protein of interest x⃗0 is compared with the mean profiles for mitochondrial 〈x⃗1〉 and nonmitochondrial proteins 〈x⃗2〉 to determine its localization. In this example, the protein of interest is assigned to the mitochondrion because the query protein's phylogenetic profile more closely resembles the mean profile of mitochondrial proteins than the mean profile of cytosolic proteins.
Testing Functional Overlap.
Protein functions were described by using Swiss-Prot protein database keywords (13). The functions of two sets of proteins were compared by calculating the Jaccard coefficient between the keywords from the two sets and comparing that to the coefficients observed in random trials. The Jaccard coefficient (C) varies from zero to one and is the normalized overlap between two sets, defined as follows: Given two groups of proteins p1 and p2, C = (number of keywords in p1 AND p2)/(number of keywords in p1 OR p2). This was compared with C calculated from p1 and 1,000 randomly chosen groups of proteins, choosing each random group with the same number of proteins as p2.
Results
Phylogenetic Profiles Vary with Subcellular Location.
The mean phylogenetic profile of mitochondrial proteins, plotted horizontally in Fig. 1A shows the average similarity in sequence of mitochondrial proteins to homologs of various genomes. This phylogenetic profile is seen to be different from the mean phylogenetic profile of proteins destined for other locations. Notably, proteins destined for secretion and for the cytosol, peroxisomes, and plasma membrane also have reasonably distinct mean phylogenetic profiles. The observed relationships between the different phylogenetic profiles of proteins of different subcellular compartments are plotted in Fig. 1B.
Analyzing Known Mitochondrial Proteins.
The algorithm of Fig. 2 for assigning query proteins to cellular locations was tested on yeast proteins whose subcellular locations had previously been determined experimentally or predicted by homology to experimentally localized proteins [384 mitochondrial proteins, 598 cytosolic, 945 nuclear, 57 lysosomal, 66 Golgi, 189 endoplasmic reticulum, 96 cytoskeletal, 198 plasma membrane, 45 peroxisomal proteins; only 26 mitochondrial proteins were also known to be localized in other compartments (14)]. Analysis of the known mitochondrial proteins revealed that they fall into three categories: prokaryote-derived mitochondrial proteins, which have one or more homologs in the set of prokaryotes listed in Fig. 1A; eukaryote-derived mitochondrial proteins, which have no homologs in the prokaryotic genomes but have one or more homologs in the eukaryotes; and organism-specific mitochondrial proteins, which are operationally defined as lacking homologs in all genomes listed in Fig. 1A and thus have phylogenetic profiles of all 1.0s. The subcellular locations of organism-specific proteins are impossible to assign by this method and were eliminated from our analyses. The proportions of proteins in each of these three categories are shown in Table 1.
Table 1.
The numbers of yeast and worm proteins observed and predicted to target mitochondria
| Organism | Experimental
|
Estimated
total number of mitochondrial proteins*
|
|||||
|---|---|---|---|---|---|---|---|
| Prokaryote-derived† (% of total) | Eukaryote-derived‡ (% of total) | Organism-specific§ (% of total) | Prokaryote-derived† | Eukaryote-derived‡ | Organism-specific§ | Total (% of genome) | |
| Yeast | 223 (58%) | 75 (20%) | 86 (22%) | 365 ± 28 | 126 ± 10 | 138 ± 1 | 630 ± 49 (10%) |
| Worm | 89 (61%) | 55 (38%) | 1 (1%) | 405 ± 85 | 252 ± 53 | 7 ± 1 | 663 ± 139 (4%) |
Predicted mitochondrial proteins are proteins from the complete genomes of yeast or worm assigned to mitochondria by using the algorithm of Fig. 2 with a training set consisting of all experimentally localized yeast proteins.
Values shown are averages of predictions for each of the 60 values of coverage and accuracy plotted in Fig. 3, scaled up as described in the text.
Proteins with ≥1 homolog detected in the prokaryotic genomes listed in Fig. 1A.
Proteins with no homologs in the prokaryotic genomes in Fig. 1A, but with eukaryotic homologs.
Proteins with no homologs detected in genomes listed in Fig. 1A.
Testing the Algorithm for Predicting Mitochondrial Proteins.
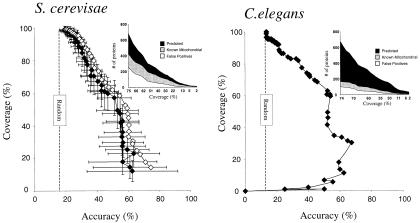
Four tests of the assignment algorithm of Fig. 2 were applied: First, we predicted the location of yeast proteins of known localization. Results of this self-consistency test are plotted by open diamonds in Fig. 3. Second, to remove bias introduced by testing the algorithm on the training set, a jackknife test was performed, withholding randomly chosen proteins from the training set and testing the algorithm on the withheld proteins, repeating the test 100 times with different random sets. Third, the algorithm was trained on yeast proteins and tested on experimentally localized worm proteins (ref. 14, 145 mitochondrial proteins and 1,649 nonmitochondrial proteins). Results from these jackknife tests are shown by filled diamonds in Fig. 3. From the jackknife test, yeast proteins can be correctly localized to mitochondria with a coverage of 58% at 50% accuracy. An accuracy of assignment of 50% is far above random (17% as shown in Fig. 3), even though we are making a binary choice of mitochondrial protein or nonmitochondrial protein. The reason is that the number of mitochondrial proteins is far smaller than the number of nonmitochondrial proteins, accounting for ≈17% of the training set. Using yeast proteins as the training set and predicting localization of experimentally localized worm proteins shows that 65% of worm nucleus-encoded mitochondrial proteins can be localized at 50% accuracy.
Figure 3.
Assignment of nuclear genome-encoded proteins to mitochondria. (Left) For yeast, a jackknife (♦ with error bars indicating ±1 SD) test on experimentally localized yeast proteins showing the method coverage (fraction of mitochondrial proteins correctly assigned) plotted versus the method accuracy (fraction of proteins assigned to mitochondria known to be mitochondrial). For comparison, results of a self-consistency test (⋄) are overlaid. (Inset) The (noncumulative) number of known (gray curve) and newly predicted (black curve) mitochondrial proteins for each coverage level, along with the number of known false positive predictions (white curve). One hundred jackknife trials were performed, randomly removing 10% of the proteins for each trial. The performance of a completely random classifier is shown as a vertical dashed line. (Right) Predicted localization of experimentally localized worm proteins by using yeast proteins as the training set. Axes are as in Left. (Inset) The number of worm proteins predicted to be mitochondrial, displayed as in Left. In both Left and Right, differing coverage and accuracy values were generated by varying the discrimination threshold (Δt) as described in Methods.
As an added check on our method, we asked whether the functions of the newly predicted mitochondrial proteins match the functions of the known mitochondrial proteins better than the function of randomly chosen sets of proteins. To measure this functional similarity, we calculated the Jaccard coefficient between the annotation (13) of predicted and known mitochondrial proteins (26.1% for proteins localized with 75% accuracy in the self-consistency test) and compared this to 1,000 random trials (17.9% ± 3.5%). The annotation of the predicted mitochondrial proteins matched the annotation of the known mitochondrial proteins much better (2.3σ) than the annotation of randomly chosen proteins, implying that the predicted mitochondrial proteins have functions consistent with known mitochondrial proteins. In contrast, the function of cytosolic proteins matches the predicted mitochondrial proteins about as well (0.1σ) as the annotation of randomly chosen proteins, reflecting the fact that random sets are predominantly composed of cytosolic proteins.
Applying the Algorithm to the Genomes of Yeast and Worm.
Applying the algorithm of Fig. 2 on all yeast proteins (9) with homologs in the genomes in Fig. 1A allowed the assignment of 361 proteins to the mitochondrion with 50% confidence. Multiplying the number of proteins assigned to mitochondria by the method accuracy and dividing by the coverage, where accuracy and coverage were determined in the self-consistency test, allows us to estimate the number of prokaryote-derived mitochondrial genes. Averaging the results from this calculation over all measured accuracies shows that the number in yeast is ≈370 genes. From the number of prokaryote-derived mitochondrial genes and their known proportion (58%) in the experimental mitochondrial protein set, we estimate there are ≈630 total mitochondrion-targeted genes in yeast, or 10% of the genome. Performing a similar analysis on worm reveals that worm contains an estimated 400 prokaryote-derived mitochondrial genes, scaling up to a total of ≈660 mitochondrion-targeted proteins, or 4% of the genome. The number of known and predicted mitochondrial genes and their inferred evolutionary origins are summarized in Table 1.
Of the experimentally localized mitochondrial proteins, we find 40% have detectable amino-terminal signal peptides (15). As summarized in Table 2, this proportion is much lower (9%) in our newly predicted mitochondrial proteins. Of the proteins either experimentally localized to mitochondria (14) or predicted here at >50% accuracy, 25% have signal peptides.
Table 2.
Fraction of yeast mitochondrial proteins with predicted transmembrane segments or signal peptides
| Structure | Fraction (%) with structure
|
||||||
|---|---|---|---|---|---|---|---|
| Experimentally localized
proteins
|
Predicted mitochondrial proteins*
|
||||||
| Prokaryote-derived† | Eukaryote-derived‡ | Organism-specific§ | All | Newly predicted | Predicted and known¶ | All predicted | |
| Transmembrane segments‖ | 36 | 55 | 27 | 37 | 46 ± 5 | 32 ± 4 | 40 ± 3 |
| Signal peptides** | 53 | 15 | 28 | 40 | 9 ± 2 | 53 ± 1 | 25 ± 2 |
Averages of 15 values from predictions made using predictive accuracies ranging from 50% to 75%. Each average is tabulated ± 1 SD.
Proteins with ≥1 homolog detected in the prokaryotic genomes listed in Fig. 1A.
Proteins with no homologs in the prokaryotic genomes in Fig. 1A, but with eukaryotic homologs.
Proteins with no homologs detected in genomes listed in Fig. 1A.
True positive predictions.
Fractions were calculated as the proteins with at least one predicted transmembrane segment, calculated as in Klein et al. (27) and implemented in psort-ii (15).
Fractions were calculated as proteins with a score >0 from the mitdisc mitochondrial targeting signal predictor of psort-ii (15).
The Function of Yeast Mitochondrial Proteins.
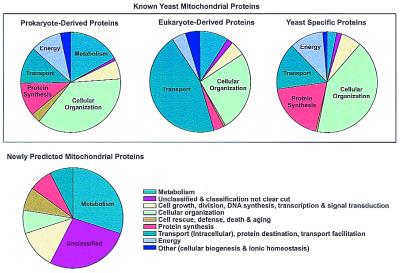
The functions of known and newly predicted yeast mitochondrial proteins are shown in Fig. 4. Notably, known mitochondrial genes have different general functions depending on their phylogenetic origins: prokaryote-derived mitochondrial proteins predominantly perform roles in metabolism, energy production, protein synthesis, and organization of mitochondria. By contrast, almost half of the eukaryote-derived mitochondrial proteins are devoted to transport and protein targeting; a significant portion are also involved in metabolism and mitochondrial organization. The yeast-specific mitochondrial proteins are predominantly involved in protein synthesis and mitochondrial organization, but also have roles in transport, transcription, metabolism, and energy production.
Figure 4.
Functions of yeast mitochondrial proteins are plotted for known mitochondrial proteins (upper three pie charts) and for the newly predicted mitochondrial proteins (lower pie chart). Each pie chart shows the percentage of proteins with a given function. Known mitochondrial proteins can be operationally divided into three populations: those with homologs in eubacteria or archaea (prokaryote-derived mitochondrial proteins), those with homologs only in other eukaryotes (eukaryote-derived mitochondrial proteins), and those without detectable homologs in the set of complete genomes (organism-specific mitochondrial proteins). Many functional systems, such as the mitochondrial ribosome, have components from more than one category of genes. The organism-specific mitochondrial proteins may be conserved in related species; many of the yeast-specific genes are conserved in other fungi as well, although absent in the more distantly related eukaryotes listed in Fig. 1A. Functional categories are defined as in the MIPS (Munich Information Center for Protein Sequences) database (29). For this analysis, mitochondrial proteins were predicted with an accuracy of 70% as scored by the self-consistency test.
Although our prediction accuracy is imperfect, we have examined the functions of yeast proteins of unknown localization but that we assign to mitochondria. The yeast proteins newly predicted to target the mitochondrion (Fig. 4) are predominantly involved in metabolism and mitochondrial organization, but they include many new proteins with functions in mitochondrial growth and division, general stress response, and a significant portion of proteins (21–28%, depending on prediction accuracy) of entirely unknown function. Among the proteins predicted to target mitochondria (available at http://www.doe-mbi.ucla.edu) are several proteins homologous to bacterial antibiotic transporters (YBR293W, YHR048W, YOR273C, and YPR156C), as well as other transporters (YGR224W). Also predicted to be mitochondrial are proteins involved in tetrahydrobiopterin and folate synthesis (FOL1 and FOL2), porphyrin synthesis (YNR029C), various other metabolisms, such as methionine/threonine synthesis (THR2 and THR3), and heme-related metabolism [the HemK homolog YNL063W and the hemolysin homolog YOL060C, whose knockout produces aberrant mitochondria (16)]. Proteins of entirely unknown function predicted to target mitochondria are listed below; each protein is referred to by the name of the encoding gene in the yeast genome (9): YBL060W, YCR059C, YDL001W, YDL201W, YDR196C, YDR282C, YDR336W, YDR539W, YER057C, YFR006W, YFR048W, YGR021W, YIL051C, YIL145C, YIR042C, YJL060W, YKR087C, YLL027W, YLR401C, YLR405W, YLR426W, YML080W, YMR278W, YMR293C, YNL026W, YNR015W, YOL008W, YOL071W, YOR111W, YOR246C, and YPL017C.
Subcellular Locations of Some Proteins Are Consistently Mispredicted.
When we examine false positive predictions, we observe some proteins, such as the β-oxidation and fatty-acid transport proteins PXA1, PXA2, FOX2, and FAS1, whose phylogenetic profiles match mitochondrial phylogenetic profiles extremely well, suggesting mitochondrial ancestry, but are known to be targeted to the peroxisome or cytosol (17–19).
Discussion
We show that the phylogenetic profile is a useful tool for assigning subcellular localization. The observed relationships between the phylogenetic profiles are doubtless a consequence of differential phylogeny of some of these organelles, such as the endosymbiotic origins of mitochondria (1, 2), and possibly peroxisomes (20) and the nucleus, suggested to be derived from eocytes (21).
Two general results come out of this analysis: (i) the number of nucleus-encoded mitochondrial genes is ≈650, accounting for ≈10% of the yeast genome and 4% of the worm genome. Of these genes, ≈370 are conserved among microorganisms, providing an estimate of the number of genes contributed by the ancestral mitochondrial genome. (ii) Eukaryotic cells have many eukaryote-derived genes that are transported into mitochondria. The large number of genes predicted to be associated with mitochondria (roughly constant between yeast and worm and accounting for 4–10% of the nuclear genome) and the many eukaryote-derived mitochondrial genes support the idea that after the endosymbiosis of the mitochondrial progenitor, the function of mitochondria continued to evolve, as encoded by both mitochondrial and nuclear genomes. The relatively large fraction of these proteins (22%) specific to yeast raises the possibility that mitochondria may have specialized functions depending on the host cell.
The Number of Mitochondrial Proteins.
Our estimate of 600–700 mitochondrial proteins is supported by two-dimensional gel electrophoresis (2D gel) of the mitochondrial proteome. Examination of 2D gel data derived from whole rat liver mitochondria and human placental mitochondria reveals ≈250–350 visible proteins (22, 23). Given the limited efficiency of observing a protein by 2D gel analysis and the chance that any particular protein is expressed at a particular time, we know that the actual number of mitochondrial proteins will be significantly larger. 2D gel analyses of whole yeast cells and the soluble cell fractions (Lundberg laboratory, http://yeast-2dpage.gmm.gu.se/; ref. 24) reveal a wide range (700–1,300) of proteins, 13–36% of the proteins expected from the genome sequence of yeast (9). Likewise, whole cell 2D gel analysis of Escherichia coli (25) reveals ≈1,846 proteins, 43% of the 4,289 proteins in E. coli (26). Therefore, the percentage of proteins revealed on a 2D gel is ≈13–43%, giving 300 divided by 0.13 to 0.43, or about 750–2,300 proteins expected in mitochondria, of the same order of magnitude as we find in this analysis.
The relatively low fraction of signal peptides (25%) in proteins either known or predicted to target mitochondria suggests that the known amino-terminal targeting peptide-dependent transport systems account for a small portion of the proteins localized to mitochondria. One explanation for the lower fraction of proteins with signal peptides is that a relatively higher proportion of predicted mitochondrial proteins are also predicted (27) as membrane proteins (46% of newly predicted mitochondrial proteins versus 32% of experimentally localized mitochondrial proteins). Membrane proteins are known to be imported into mitochondria independently of an amino-terminal targeting signal (reviewed in ref. 28).
A Displaced Proteome?
The observation that the phylogenetic profiles of some proteins match mitochondrial profiles very well, even though the proteins are known to be localized in other compartments, raises the possibility that not all proteins of organellar ancestry are targeted back to the same organelle, and proteins may be targeted instead to other subcellular locations. These “retargeted” proteins would contribute to the average phylogenetic profiles of Fig. 1A, and therefore contribute to the observed relationships between proteins of different subcellular compartments plotted in Fig. 1B. One interpretation of these relationships, drawn in Fig. 1B superimposed over the observed tree of relationships, is that most subcellular compartments derive from the eukaryotic progenitor that hosted the mitochondrial progenitor endosymbiont, but that some proteins of mitochondrial ancestry are now targeted to the cytoplasm and plasma membrane, shifting the mean phylogenetic profile of proteins in those compartments closer to the mitochondrial profile, as suggested by the dashed arrows in Fig. 1B. These proteins may represent a displaced proteome of mitochondria, consisting of proteins of mitochondrial origin whose genes were shifted into the nuclear genome, but whose proteins were in turn localized to new compartments in the continuing evolution of the cell.
Acknowledgments
We thank Dr. Michael Thompson, Dr. Steve Wickert, Dr. Carla Koehler, and Dr. Elizabeth Neufeld for suggestions, Dr. Kenta Nakai for generously providing psort-ii, the National Institutes of Health and the Department of Energy for support, and the Swiss National Fund for a fellowship to I.X.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220399497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220399497
References
- 1.Margulis L. Origin of Eukaryotic Cells. New Haven, CT: Yale Univ. Press; 1970. [Google Scholar]
- 2.Gray M W. Curr Opin Genet Dev. 1999;9:678–687. doi: 10.1016/s0959-437x(99)00030-1. [DOI] [PubMed] [Google Scholar]
- 3.Gray M W, Burger G, Lang B F. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 4.Marcotte E M, Pellegrini M, Yeates T O, Eisenberg D. J Mol Biol. 1999;293:151–160. doi: 10.1006/jmbi.1999.3136. [DOI] [PubMed] [Google Scholar]
- 5.Chou K C, Elrod D W. Protein Eng. 1999;12:107–118. doi: 10.1093/protein/12.2.107. [DOI] [PubMed] [Google Scholar]
- 6.Tatusov R L, Galperin M Y, Natale D A, Koonin E V. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ragan M A, Gaasterland T. Microb Comp Genomics. 1998;3:219–235. doi: 10.1089/omi.1.1998.3.219. [DOI] [PubMed] [Google Scholar]
- 8.Pellegrini M, Marcotte E M, Thompson M J, Eisenberg D, Yeates T O. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 10.C. elegans Genome Sequencing Consortium. Science. 1998;282:2012–2018. [Google Scholar]
- 11.Marcotte E M. Curr Opin Struct Biol. 2000;10:359–365. doi: 10.1016/s0959-440x(00)00097-x. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bairoch A, Apweiler R. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costanzo M C, Hogan J D, Cusick M E, Davis B P, Fancher A M, Hodges P E, Kondu P, Lengieza C, Lew-Smith J E, Lingner C, et al. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakai K, Horton P. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 16.Entian K D, Schuster T, Hegemann J H, Becher D, Feldmann H, Guldener U, Gotz R, Hansen M, Hollenberg C P, Jansen G, et al. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- 17.Shani N, Valle D. Proc Natl Acad Sci USA. 1996;93:11901–11906. doi: 10.1073/pnas.93.21.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweizer E, Werkmeister K, Jain M K. Mol Cell Biochem. 1978;21:95–107. doi: 10.1007/BF00240280. [DOI] [PubMed] [Google Scholar]
- 19.Qin Y M, Marttila M S, Haapalainen A M, Siivari K M, Glumoff T, Hiltunen J K. J Biol Chem. 1999;274:28619–28625. doi: 10.1074/jbc.274.40.28619. [DOI] [PubMed] [Google Scholar]
- 20.de Duve C. Sci Am. 1996;274(4):50–57. doi: 10.1038/scientificamerican0496-50. [DOI] [PubMed] [Google Scholar]
- 21.Lake J A. Nature (London) 1988;331:184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- 22.Pavlica R J, Hesler C B, Lipfert L, Hirshfield I N, Haldar D. Biochim Biophys Acta. 1990;1022:115–125. doi: 10.1016/0005-2736(90)90407-f. [DOI] [PubMed] [Google Scholar]
- 23.Rabilloud T, Kieffer S, Procaccio V, Louwagie M, Courchesne P L, Patterson S D, Martinez P, Garin J, Lunardi J. Electrophoresis. 1998;19:1006–1014. doi: 10.1002/elps.1150190616. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez J C, Golaz O, Frutiger S, Schaller D, Appel R D, Bairoch A, Hughes G J, Hochstrasser D F. Electrophoresis. 1996;17:556–565. doi: 10.1002/elps.1150170326. [DOI] [PubMed] [Google Scholar]
- 25.Tonella L, Walsh B J, Sanchez J C, Ou K, Wilkins M R, Tyler M, Frutiger S, Gooley A A, Pescaru I, Appel R D, et al. Electrophoresis. 1998;19:1960–1971. doi: 10.1002/elps.1150191114. [DOI] [PubMed] [Google Scholar]
- 26.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 27.Klein P, Kanehisa M, DeLisi C. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann J M, Neupert W. Curr Opin Microbiol. 2000;3:210–214. doi: 10.1016/s1369-5274(00)00077-1. [DOI] [PubMed] [Google Scholar]
- 29.Mewes H W, Frishman D, Gruber C, Geier B, Haase D, Kaps A, Lemcke K, Mannhaupt G, Pfeiffer F, Schuller C, et al. Nucleic Acids Res. 2000;28:37–40. doi: 10.1093/nar/28.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]