Abstract
During zebrafish development, zygotic gene expression initiated at the midblastula transition converts maternal information on embryo polarity into a transcriptional read-out. Expression of a homeobox gene, vega1, is activated at midblastula transition in all blastomeres, but is down-regulated dorsally before gastrulation. Ubiquitous expression of vega1 is maintained in bozozok mutants, in which the dorsal-specific homeobox gene bozozok/dharma (boz/dha) is disrupted and organizer formation is impaired. Vega1 inhibits expression of boz/dha and organizer-specific genes, and causes ventralization resulting in a headless phenotype. In contrast, VP16-vega1, a fusion including the Vega1 homeodomain and VP16 activation domain, elicits ectopic expression of organizer genes and suppresses several aspects of the boz mutant phenotype. We propose that boz/dha-dependent down-regulation of vega1 in the dorsal region is an early essential step in organizer formation in zebrafish.
Recent studies in Xenopus suggested that transcriptional events between the midblastula transition (MBT) and the beginning of gastrulation establish embryonic polarity and axis formation (1–4). Maternal β-catenin signaling at the dorsal side activates the homeobox genes siamois and twin immediately after MBT. Siamois is a transcriptional activator that positively regulates the expression of Spemann organizer genes and can induce a complete secondary axis (3, 4). In contrast, Vent family homeobox genes, such as Xvent-1, function as transcriptional repressors and appear to be activated by bone morphogenetic protein (BMP) signaling only ventrally (5–8). Although Vent genes inhibit organizer formation, these genes do not regulate siamois expression (8).
Recently, bozozok/dharma (boz/dha; also known as nieuwkoid) was identified in zebrafish as a functional mediator of the maternal β-catenin signal on the dorsal side of the blastula (9–11). Zygotic boz/dha expression arises immediately after MBT in a small group of dorsal blastomeres. By the dome stage, boz/dha expression becomes progressively restricted to the dorsal yolk syncytial layer (YSL), an extraembryonic structure formed by fusion of marginal blastomeres with the underlying yolk cell. The dorsal YSL appears to possess Nieuwkoop center-like activity (12), contributing to the establishment of the organizer in a non-cell autonomous manner. Misexpression of boz/dha can induce the organizer gene goosecoid (gsc) in a non-cell autonomous manner (9, 10). Furthermore, boz mutants form an incomplete organizer and exhibit dorso-anterior deficiencies, indicating that boz/dha is a key component of gastrula organizer formation (11, 13, 14).
Here, we describe a homeobox gene vega1, isolated by using a subtracted library enriched in zygotically activated genes. We demonstrate that vega1 has a mutually antagonistic relationship with boz/dha and an apparent critical role in early dorso-ventral and antero-posterior patterning of the zebrafish embryo between MBT and organizer formation.
Materials and Methods
Isolation of Vega1.
Total RNA of early stage (0–32 cell stage) and dome stage (about 1.5 h after MBT) zebrafish embryos was extracted by using Trizol reagent (GIBCO/BRL), and poly(A)+ RNA was isolated by using the Fast Track Kit (Invitrogen). The early stage cDNA library was subtracted from the dome stage cDNA library, using the PCR-selected cDNA Subtraction Kit (CLONTECH) as described previously (15). cDNAs were selected by further screening using whole-mount in situ hybridization. One gene, which we named vega1, encodes a homeodomain protein. Full-length vega1 clones were isolated by rapid amplification of cDNA ends (RACE), using the Marathon cDNA Amplification Kit (CLONTECH). The nucleotide sequence of vega1 was obtained by sequencing five independent clones. The GenBank accession number of vega1 is AF193837.
Construction.
The coding region of Vega1 (amino acids 1–242) was inserted into pCS2 + vector. Vega1-HA (amino acids 1–242), Vega1-ΔN (amino acids 48–242), ENG-Vega1 (Drosophila Engrailed, amino acids 1–298, plus Vega1, amino acids 100–242), and VP16-Vega1 (Herpes simplex virus I VP16 gene, amino acids 410–490, plus Vega1, amino acids 101–242) were fused to the HA epitope at the C terminus and inserted into the pCS2 + vector. The HA epitope was fused to Vega1-ΔHD (amino acids 1–116) at the N terminus and inserted into the pCS2 + vector. The sequence of inserted cDNAs and their junctions with the backbone were confirmed for all constructs by sequencing.
Dual Luciferase Assay.
For luciferase assay experiments, COS-7 cells (0.75 × 105) were plated in a 24-well dish 12 h before transfection. Cells were transfected with total 2 μg of plasmid DNA (0.4 or 1.2 μg of indicated expression vector plus 1.2 or 1.6 μg of pCS2 + vector, 398 ng of p4.0gsclux, and 2 ng of pRL-SV40) and GenePORTER transfection reagent (10 μl) according to manufacturer's directions (Gene Therapy Systems). The dual luciferase assay system (Promega) was used. Twenty-4 h after transfection, cells were harvested and incubated with lysis buffer (100 μl), and aliquots of 20 μl were used for luciferase assays according to the manufacturer's instructions.
Results
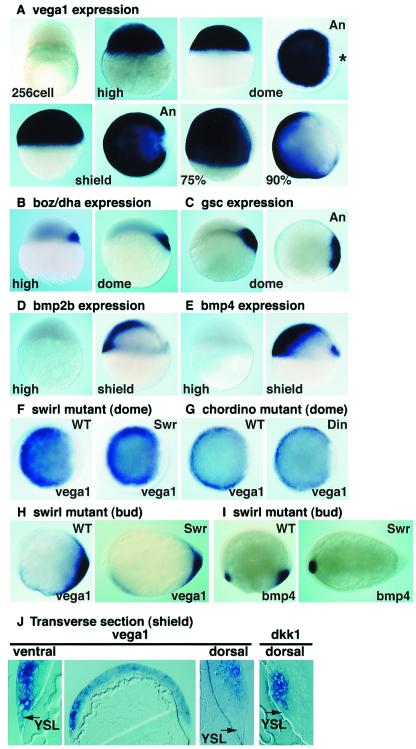
Vega1 is not expressed maternally and is activated immediately after MBT in all blastomeres as seen by in situ hybridization (Fig. 1A) and Northern blotting (data not shown). About 1 h later (dome stage), vega1 transcripts are no longer detected in a small dorsal domain including blastomeres and YSL (Fig. 1A, ∗). Dorsal exclusion of vega1 transcripts continues during early gastrulation in the shield, the equivalent of the Spemann organizer, and is maintained throughout gastrulation (Fig. 1A). Both dorsal YSL and the adjacent deep blastomeres are vega1 negative, whereas ventral YSL and blastoderm are strongly stained (Fig. 1J). We compared vega1 expression to that of dorsal specific homeobox genes boz/dha and gsc. At the high stage, ubiquitous vega1 expression overlaps the early boz/dha expression in a few dorsal blastomeres (Fig. 1B) (9–11). With the exclusion of vega1 from a dorsal region at dome stage, its expression becomes complementary to that of boz/dha and gsc, although regions of overlapping expression remain (Fig. 1 A–C).
Figure 1.
Vega1 is activated at the MBT and expressed in a complementary pattern to the dorsal genes, boz/dha and gsc. Whole-mount in situ hybridization of vega1 (A, F–H), boz/dha (B), gsc (C), bmp2b (D), and bmp4 (E and I). (A–G) Dorsal is to the right where known; An, animal view; 75% and 90%, 75% and 90% epiboly stage. Asterisk in dome stage animal view points to vega1-depleted region. (H–I) Anterior is to the left, dorsal up. (F and G) Expression of vega1 is not changed in swirltc300/tc300 and chordinott250/tt250 mutant embryos at dome stage. Genotyping of mutant embryos was performed by PCR. (H and I) At bud stage, vega1 is partly down-regulated in swirltc300/tc300 mutant embryos, whereas bmp4 expression in the posterior region is abolished. (J) Transverse section of shield stage embryo stained by in situ hybridization for vega1 or dkk1. It is noteworthy that the dkk1 expression domain is vega1 negative. Arrows indicate yolk syncytial layer.
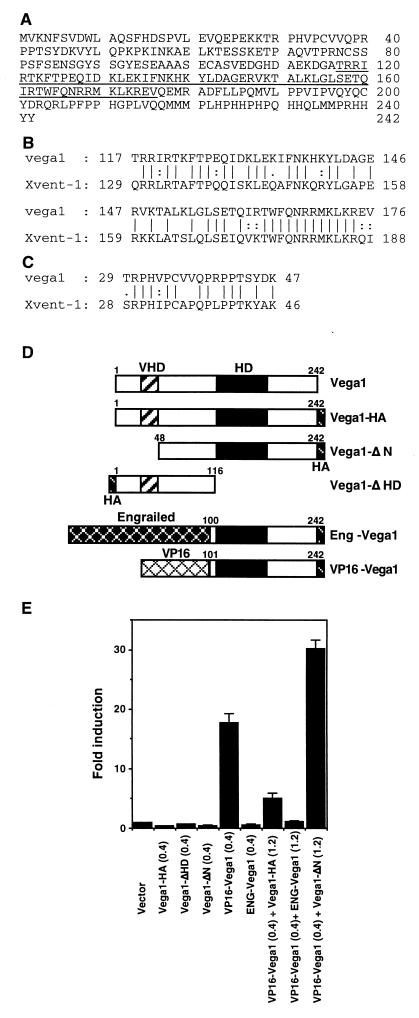
Vega1 encodes a homeodomain protein of 242 amino acids, with 65% sequence identity between its homeodomain and that of Xenopus Xvent-1, the most similar protein in the database (Fig. 2 A and B). A second region of high similarity is named the Vent-1 Homology Domain (VHD) (Fig. 2C), whereas the overall sequence identity between Vega1 and any Vent family protein is less than 34%. Because vega1 functions parallel or upstream of BMP (see below), vega1 does not appear to be the zebrafish ortholog of Xvent-1 or of any other reported Vent family gene (16).
Figure 2.
Vega1 encodes a homeodomain protein that can regulate the goosecoid promoter. (A) Predicted amino acid sequence of the Vega1 protein with the homeodomain underlined. (B) Sequence alignment of the homeodomains of Vega1 and Xvent-1. Bars and dots mean identical amino acid and similar amino acid, respectively. (C) Comparison of N-terminal Vent-1 homology domain (VHD) of Vega1 and Xvent-1. (D) Schematic representation of Vega1 mutant constructs. HD and HA indicate homeodomain and influenza hemagglutinin epitope, respectively. (E) Regulation of the gsc promoter by Vega1 constructs. COS-7 cells were transfected with the indicated cDNA in addition to reporter (p4.0gsclux) and control plasmids (pRL-SV40). Luciferase activity was normalized by the internal control plasmid and expressed relative to the activity of the vector alone. The error bars represent the standard deviation of three experiments. Protein expression levels of Vega1 constructs were comparable as judged by anti-HA Western blotting.
We hypothesized that vega1 directly suppresses gsc expression ventrally and laterally, and regulates the location and timing of its dorsal expression, because gsc expression starts as vega1 is down-regulated dorsally. To examine this possibility, we used a zebrafish gsc/luciferase reporter construct and fusion proteins between the Vega1 DNA binding domain and the previously characterized activation or repressor domains of VP16 and Engrailed, respectively (Fig. 2D). Vega1 constructs were cotransfected with p4.0gsclux plasmid (4.0 kb zebrafish gsc upstream region linked to luciferase) (17) and internal control plasmid (pRL-SV40) into COS-7 cells. As shown in Fig. 2E, vega1, truncated constructs, or the Engrailed fusion failed to stimulate the gsc promoter; in contrast, VP16-vega1 strongly activated this promoter. Interestingly, this VP16-vega1-induced gsc promoter activation was inhibited by cotransfection of either vega1-HA or ENG-vega1, whereas vega1-ΔN, which still carries the homeodomain, did not inhibit the stimulation. Furthermore, analysis of deletion mutants of the gsc promoter showed that a 1-kb segment (from −1711 to −738) is required for VP16-vega1 responsiveness, and wild-type Vega1 protein, but not Vega1-ΔHD, had binding activity for this fragment (data not shown). These results suggest that Vega1 functions as a transcriptional repressor for the gsc promoter, and that the N-terminal domain (residues 1–47) of Vega1 is required for repressor activity. Xvent-1 is known to act as a transcriptional repressor for the XFD-1/XFKH1/pintallavis promoter by its N-terminal domain (18), which contains a region of high similarity between Vega1 and Xvent-1; this region, named the VHD, may constitute a new type of repressor domain.
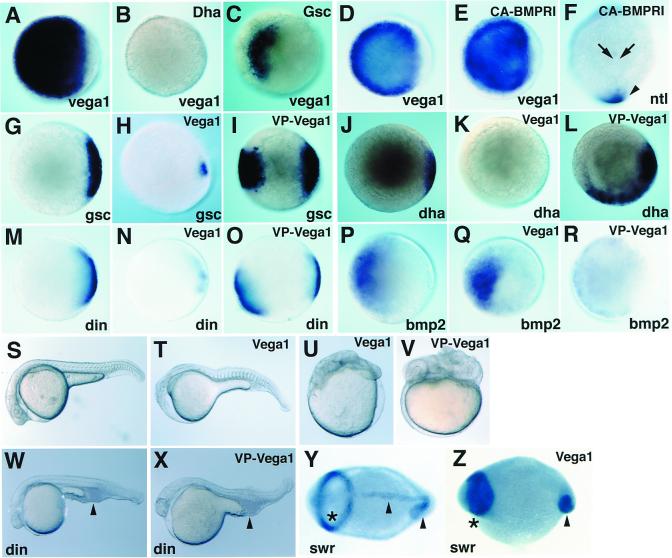
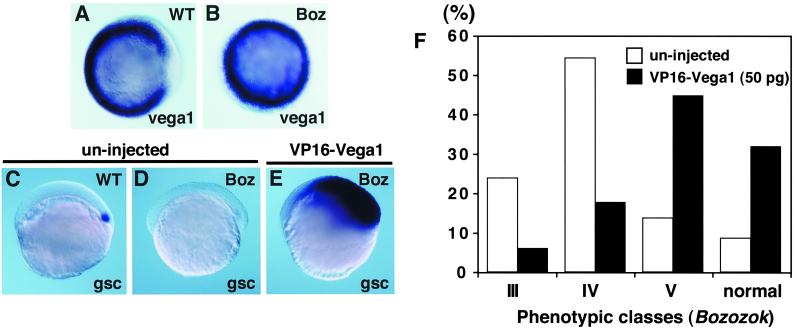
We asked how vega1 expression is suppressed in a small dorsal domain before the dome stage. The dorsal domain exhibits nuclear localization of β-catenin, which leads to the activation of dorsal genes including boz/dha and subsequently gsc, during blastula stages (11, 19). Injection of boz/dha or gsc mRNA inhibited vega1 expression at the dome stage, with boz/dha being more effective (Fig. 3 A–C). These results suggest that boz/dha and to a lesser extent, gsc, mediate suppression of vega1 at the dorsal side of the embryo during blastula stages. To test whether boz/dha is required for down-regulation of vega1 transcripts dorsally, we analyzed expression of vega1 in bozm168 mutant embryos (11, 13). We found that ubiquitous vega1 expression was maintained in boz mutants through the shield stage (Fig. 4 A and B), whereas gsc expression was strongly reduced (data not shown) (11). These results indicate that zygotic boz function is required for the dorsal repression of vega1.
Figure 3.
Regulation of vega1 expression and analysis of vega1 function. Whole-mount in situ hybridization for vega1 (A–E), ntl (F), gsc (G–I), boz/dha (J–L), chordin (M–O), and bmp2b (P–R); animal views (A–E, G–R), dorsal view (F), dome stage (A–E, G–I, M–O), 30% epiboly stage (P–R), shield stage (J–-L), bud stage (F). (A–C) boz/dha mRNA (B; 25pg), gsc mRNA (C; 50pg) were injected into one to two cell stage embryos; (A) uninjected control. (D–F) RNA injections; CA-BMPRI mRNA (E and F; 200 pg) was injected into one to two cell stage embryos; (D) uninjected control. (E) Vega1 expression was unchanged. (F) Expression of ntl in the chordamesoderm region was suppressed (arrows), but not affected in the tailbud domain (arrowhead). (G–R) vega1 mRNA (H; 250 pg; and K, N, and Q, 100 pg) and VP16-vega1 mRNA (I, L, O, and R, 50 pg) were injected into one to two cell and two to eight cell stage embryos, respectively. (G, J, M, and P) Uninjected controls. (S–V) Phenotypic effects; lateral views, prim-6 (25 hpf) stage. Embryos were injected with vega1 mRNA (T and U; 100pg) or VP16-vega1 mRNA (V; 50pg) at the one to two cell stage. (S) Uninjected control. (W and X) Embryos from a dintt250/+X dintt250/+ cross were injected with VP16-vega1 mRNA (X; 100pg) at one to two cell stage. VP16-vega1 could not restore the ventral tail phenotype (arrowhead), whereas wild-type embryos injected with VP16-vega1 were dorsalized (V). (W) Uninjected dintt250/tt250 mutant. (Y and Z) Whole-mount in situ hybridization for pax2.1 plus ntl; 5-somite stage, dorsal view. Asterisk and arrowhead show pax2.1 and ntl staining domain, respectively. (Z) Embryos from a swirltc300/+X swirltc300/+ cross were injected with vega1 mRNA (150pg) at one to two cell stage. (Y) Uninjected swirltc300/tc300 mutant.
Figure 4.
VP16-vega1 can rescue both gsc expression and boz phenotype. Whole-mount in situ hybridization for vega1 (A and B, shield stage, animal view) and gsc (C–E, 40% epiboly stage, lateral view). Genotyping of boz mutants was carried out by restriction fragment polymorphism (11). (A) bozm168/+ phenotypically wild type. (B) bozm168/m168 mutant. (C) Uninjected wild-type embryo. (D) Uninjected bozm168/m168 embryo. Expression of gsc was suppressed in all uninjected boz embryos (n = 15). (E) VP16-vega1 mRNA (100pg) was injected into one-cell stage of embryos obtained from a bozm168/m168 × bozm168/m168 cross. Expansion of gsc was observed in all VP16-vega1-injected boz embryos (100%, n = 39). (F) Suppression of boz phenotype by VP16-vega1 injection. VP16-vega1 mRNA (50 pg) was injected into one-cell stage of embryos obtained from bozm168/m168 × bozm168/m168 crosses. The boz phenotype was classified by morphological criteria at 30 hpf as described previously (11). The number of uninjected and VP16-vega1-injected embryos was 59 and 85, respectively. Class V has a small break in trunk notochord with normal head structure; there is a two-to-several somite wide gap in the trunk notochord of class IV; class III shows partial cyclopia with anterior head deficiency and usually a large gap in the trunk notochord. Essentially identical results were obtained in several independent experiments.
In Xenopus, Vent family genes such as Xvent-1 function as downstream mediators of BMP signaling (5). Therefore, we examined the functional interaction between vega1 and BMP signaling. When vega1 is expressed in all blastomeres at the high stage, bmp2b expression is just starting and bmp4 expression is not detectable (Fig. 1 D and E). At the shield stage, the expression patterns of vega1 and of bmp2b/bmp4 which form a ventral-to-dorsal gradient, are quite different (Fig. 1 A, D, and E). Consistently, expression of vega1 was not affected at the mid-blastula stage in swirltc300 (swr) and chordinott250 (din) mutant embryos (Fig. 1 F and G); swr disrupts the bmp2b gene, din disrupts the BMP antagonist, Chordin (20–23). Furthermore, injection of the constitutively active form of the BMP type I receptor, BMP-RIA (24), did not cause expansion of the vega1 domain, whereas no tail (ntl) expression in the chordamesoderm was strongly suppressed (Fig. 3 D–F). These results suggest that expression of vega1 is not dependent on BMP signaling during blastula stages. At the end of gastrulation, vega1 expression was only partly suppressed in the swirl mutant, whereas bmp4 expression was abolished in the posterior domain (Fig. 1 H and I). Thus, vega1 expression during gastrulation is regulated by other signaling pathway(s) with only a small contribution from the BMP pathway.
Functional activity of vega1 and VP16-vega was examined by phenotypic changes elicited by injection of synthetic mRNA into zebrafish and Xenopus embryos. In zebrafish, both vega1 and ENG-vega1 caused ventralization with loss of anterior head structures and tail and axis defects in 53% (n = 44) (Fig. 3 T and U) and 63% (n = 49) of embryos, respectively. Likewise, vega1 mRNA injection into the two dorsal blastomeres of four-cell Xenopus embryos led to a head-less phenotype (74%; n = 66), whereas ventral injection had no effect (data not shown). In contrast, expression of VP16-vega1 in zebrafish caused a dorsalized phenotype (62%; n = 34) (Fig. 3V), similar to the phenotype obtained by ectopic boz/dha expression (9). Secondary axis formation was often observed when VP16-vega1 was injected into the two ventral blastomeres of four-cell Xenopus embryos (67%; n = 55) (data not shown).
The molecular pathways influenced by vega1 or VP16-vega1 injection were analyzed by the use of marker genes. Injection of vega1 mRNA into zebrafish embryos inhibited gsc, boz/dha, and chordin (din) expression (Fig. 3 H, K, and N). Conversely, ectopic expression of these dorsal genes was detected in VP16-vega1 injected embryos (Fig. 3 I, L, and O), whereas bmp2b expression was inhibited (Fig. 3R). Furthermore, VP16-vega1 injections resulted in expansion of the expression domains of the dorsal genes dkk1, floating head (flh), squint (sqt) and cyclops (cyc), whereas gata2 expression was inhibited (data not shown). Conversely, the expression of these genes except for gata2 was strongly suppressed by vega1 injection. Thus, vega1 acts as a ventralizing factor suppressing dorsal genes whereas VP16-vega1, an apparent antimorphic form, acts as a dorsalizing factor. Because BMP signaling is critical for maintaining ventral identity in the embryo, we examined the effects of vega1 and VP16-vega1 on swr (dorsalized) and din (ventralized) mutants. Injection of vega1 could not suppress the expansion of neuroectoderm in the swr mutant, but resulted in a loss of notochord and an anterior shift of pax2.1 expression domain (n = 129) (Fig. 3 Y and Z). This phenotype is similar to that obtained by overexpression of Noggin or Chordin in boz mutants (25), or exhibited by boz;swr double mutants (L.S.K., unpublished observation). Likewise, injection of VP16-vega1, which consistently dorsalized wild-type embryos, could not rescue the ventralized phenotye of din mutant embryos as judged by the shape of the ventral tail (Fig. 3 W and X, arrowhead). These results indicate that vega1 functions parallel or upstream of BMP signaling in axis formation in zebrafish.
Because boz/dha and vega1 are the earliest-expressed zygotic genes implicated in axis formation, we examined their functional interactions in the embryo. Boz/dha mRNA injection into the ventral blastomeres of four-cell Xenopus embryos induced secondary axes (86%, n = 57), which was inhibited by coinjection of vega1 (7%, n = 58) (data not shown). Furthermore, we tested whether VP16-vega1 can restore gsc expression and rescue other aspects of the boz mutant phenotype. VP16-vega1 mRNA injection into boz mutant embryos at the one-cell stage strongly induced gsc expression (100%, n = 39, Fig. 4E). Forebrain and notochord deficiencies of boz mutants were largely suppressed by injection of VP16-vega1 mRNA as evidenced by morphological analysis (Fig. 4F), and by the restoration of six3 (forebrain) and ntl (notochord) expression at the 8 somite stage (data not shown). These results suggest that vega1 and boz/dha functionally antagonize each other, and that repression of vega1 is an important aspect of boz/dha function in axis formation.
Discussion
The zebrafish boz/dha and the Xenopus siamois and twin genes are functional mediators of maternal β-catenin signaling in the establishment of the gastrula organizer. In Xenopus, siamois contributes to organizer formation acting as transcriptional activator of other dorsal genes (26–28). In contrast, the N-terminal domain of Boz/Dha shares sequence similarity with a characteristic domain of the transcriptional repressors Gsc and Engrailed, the so-called Gsc-Engrailed homology (GEH) domain (10). Furthermore, a Boz/Dha homeodomain-Engrailed repressor domain fusion protein has similar dorsalizing activity as wild type Boz/Dha, suggesting that this factor functions as a transcriptional repressor (M. Hibi, Y. Yamanaka, T. Hirano, personal communication). Therefore, the mechanism of boz/dha action appears distinct from that of siamois.
In this paper, we have demonstrated an antagonistic interaction between vega1 and boz/dha that is essential for organizer formation, providing a plausible mechanism through which the repressor Boz/Dha may exercise its dorsalizing role. The rapid clearing of vega1 mRNA from the future dorsal side of the embryo immediately follows the initiation of boz/dha expression and depends on zygotic boz/dha function. This dependence on boz is specific, as no change in vega1 expression was observed in the BMP signaling mutants swr and din. Interestingly, boz/dha is also required for down-regulation of the bmp2b (14, 25), and Boz/Dha protein directly binds to the promoter region of bmp2b gene (T.C. Leung, W. Driever, personal communication), indicating that boz/dha contributes the dorsal suppression of both vega1 and bmp2b genes. We have shown that vega1 and boz/dha as well as vega1 and gsc reciprocally repress each other's expression in the blastula embryo. Therefore, the initial stage of axis formation by zygotically activated genes may be based on the balance of expression levels of these homeobox genes and involves progressive refinement of their expression domains. One attractive possibility is that both boz/dha and subsequently gsc directly bind to the vega1 promoter and repress its activity. Functional antagonism is also indicated by the ability of vega1 to inhibit the axis-inducing activity of boz/dha, whereas VP16-vega1 can largely restore marker expression and morphology in boz mutant embryos. Furthermore, analysis of zebrafish mutants with elevated BMP signaling demonstrates that vega1 functions in parallel or upstream of BMP signaling in axis specification in the zebrafish.
We also were able to show that vega1 can affect the expression of several dorsal genes. The effect of vega1 constructs on gsc reporter activity and the restoration of gsc expression in boz mutants by VP16-vega1 indicate that gsc is one of the direct targets of the Vega1 repressor. Furthermore, VP16-vega1 strongly induced the ectopic expansion of dkk1, sqt, and cyc genes that encode secreted factors involved in axis formation (29–32). Because early expression of nodal genes is not dependent on boz/dha (33), these observations suggest that vega1 may inhibit the expression of nodal genes in addition to boz/dha. In summary, we suggest that elimination of vega1 transcripts from a small dorsal domain by boz/dha is the first post-MBT step in axis formation, generating a permissive region for the expression of dorsal genes such as gsc, leading to the development of the gastrula organizer.
Acknowledgments
We thank D. J. Grunwald, L. I. Zon, D. Kessler, N. Ueno, M. Hibi, T. Hirano, and R. Harland for reagents, C.-H. Kim and members of the Dawid Laboratory for helpful discussion, and M. Tsang, L. Kodjabachian and T. Kudoh for critical reading of the manuscript. A.K. was supported by a postdoctoral fellowship for Research Abroad of the Japan Society for the Promotion of Science. Work in the L.S.K. laboratory was supported by March of Dimes Birth Defects Foundation and Pew Scholars Program in Biomedical Research.
Abbreviations
- MBT
midblastula transition
- YSL
yolk syncytial layer
- VHD
Vent-1 Homology Domain
- boz/dha
bozozok/dharma
- BMP
bone morphogenetic protein
Footnotes
Data deposition: The sequence reported in this paper has been submitted to the GenBank database (accession no. AF193837).
References
- 1.Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J. Development (Cambridge, UK) 1996;122:2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- 2.Fagotto F, Guger K, Gumbiner B M. Development (Cambridge, UK) 1997;124:453–460. doi: 10.1242/dev.124.2.453. [DOI] [PubMed] [Google Scholar]
- 3.Lemaire P, Garrett N, Gurdon J B. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 4.Kessler D S. Proc Natl Acad Sci USA. 1997;94:13017–22. doi: 10.1073/pnas.94.24.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. EMBO J. 1995;14:6268–6279. doi: 10.1002/j.1460-2075.1995.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladher R, Mohun T J, Smith J C, Snape A M. Development (Cambridge, UK) 1996;122:2385–2394. doi: 10.1242/dev.122.8.2385. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt J E, von Dassow G, Kimelman D. Development (Cambridge, UK) 1996;122:1711–1721. doi: 10.1242/dev.122.6.1711. [DOI] [PubMed] [Google Scholar]
- 8.Shapira E, Marom K, Levy V, Yelin R, Fainsod A. Mech Dev. 2000;90:77–87. doi: 10.1016/s0925-4773(99)00283-x. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka Y, Mizuno T, Sasai Y, Kishi M, Takeda H, Kim C H, Hibi M, Hirano T. Genes Dev. 1998;12:2345–2353. doi: 10.1101/gad.12.15.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koos D S, Ho R K. Curr Biol. 1998;8:1199–1206. doi: 10.1016/s0960-9822(07)00509-x. [DOI] [PubMed] [Google Scholar]
- 11.Fekany K, Yamanaka Y, Leung T, Sirotkin H I, Topczewski J, Gates M A, Hibi M, Renucci A, Stemple D, Radbill A, et al. Development (Cambridge, UK) 1999;126:1427–1438. doi: 10.1242/dev.126.7.1427. [DOI] [PubMed] [Google Scholar]
- 12.Kodjabachian L, Dawid I B, Toyama R. Dev Biol. 1999;213:231–245. doi: 10.1006/dbio.1999.9392. [DOI] [PubMed] [Google Scholar]
- 13.Solnica-Krezel L, Stemple D L, Mountcastle-Shah E, Rangini Z, Neuhauss S C, Malicki J, Schier A F, Stainier D Y, Zwartkruis F, Abdelilah S, Driever W. Development (Cambridge, UK) 1996;123:67–80. doi: 10.1242/dev.123.1.67. [DOI] [PubMed] [Google Scholar]
- 14.Koos D S, Ho R K. Dev Biol. 1999;215:190–207. doi: 10.1006/dbio.1999.9479. [DOI] [PubMed] [Google Scholar]
- 15.Kawahara, A. & Dawid, I. B. (2000) Mech. Dev., in press. [DOI] [PubMed]
- 16.Rastegar S, Friedle H, Frommer G, Knochel W. Mech Dev. 1999;81:139–149. doi: 10.1016/s0925-4773(98)00239-1. [DOI] [PubMed] [Google Scholar]
- 17.McKendry R, Harland R M, Stachel S E. Dev Biol. 1998;204:172–186. doi: 10.1006/dbio.1998.9065. [DOI] [PubMed] [Google Scholar]
- 18.Friedle H, Rastegar S, Paul H, Kaufmann E, Knochel W. EMBO J. 1998;17:2298–2307. doi: 10.1093/emboj/17.8.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stachel S E, Grunwald D J, Myers P Z. Development (Cambridge, UK) 1993;117:1261–1274. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- 20.Kishimoto Y, Lee K H, Zon L, Hammerschmidt M, Schulte-Merker S. Development (Cambridge, UK) 1997;124:4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen V H, Schmid B, Trout J, Connors S A, Ekker M, Mullins M C. Dev Biol. 1998;199:93–110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- 22.Schulte-Merker S, Lee K J, McMahon A P, Hammerschmidt M. Nature (London) 1997;387:862–863. doi: 10.1038/43092. [DOI] [PubMed] [Google Scholar]
- 23.Piccolo S, Sasai Y, Lu B, De Robertis E M. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido M, Tada M, Takeda H, Kuroiwa A, Ueno N. Development (Cambridge, UK) 1999;126:181–190. doi: 10.1242/dev.126.1.181. [DOI] [PubMed] [Google Scholar]
- 25.Fekany-Lee K, Gonzalez E, Miller-Bertoglio V, Solnica-Krezel L. Development (Cambridge, UK) 2000;127:2333–2345. doi: 10.1242/dev.127.11.2333. [DOI] [PubMed] [Google Scholar]
- 26.Lemaire P, Kodjabachian L. Trends Genet. 1996;12:525–531. doi: 10.1016/s0168-9525(97)81401-1. [DOI] [PubMed] [Google Scholar]
- 27.Harland R, Gerhart J. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 28.Moon R T, Kimelman D. BioEssays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 29.Rebagliati M R, Toyama R, Haffter P, Dawid I B. Proc Natl Acad Sci USA. 1998;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman B, Gates M A, Egan E S, Dougan S T, Rennebeck G, Sirotkin H I, Schier A F, Talbot W S. Nature (London) 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- 31.Sampath K, Rubinstein A L, Cheng A M, Liang J O, Fekany K, Solnica-Krezel L, Korzh V, Halpern M E, Wright C V. Nature (London) 1998;395:185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto H, Itoh M, Yamanaka Y, Yamashita S, Shimizu T, Solnica-Krezel L, Hibi M, Hirano T. Dev Biol. 2000;217:138–152. doi: 10.1006/dbio.1999.9537. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu T, Yamanaka Y, Ryu S, Hashimoto H, Yabe T, Hirata T, Bae Y, Hibi M, Hirano T. Mech Dev. 2000;91:293–303. doi: 10.1016/s0925-4773(99)00319-6. [DOI] [PubMed] [Google Scholar]