Abstract
Self-organization has been demonstrated in a variety of systems ranging from chemical-molecular to ecosystem levels, and evidence is accumulating that it is also fundamental for animal development. Yet, self-organization can be approached experimentally in only a few animal systems. Cells isolated from the simple metazoan Hydra can aggregate and form a complete animal by self-organization. By using this experimental system, we found that clusters of 5–15 epithelial cells are necessary and sufficient to form de novo head-organizing centers in an aggregate. Such organizers presumably arise by a community effect from a small number of cells that express the conserved HyBra1 and HyWnt genes. These local sources then act to pattern and instruct the surrounding cells as well as generate a field of lateral inhibition that ranges up to 1,000 μm. We propose that conserved patterning systems in higher animals originate from extremely robust and flexible molecular self-organizing systems that were selected for during early metazoan evolution.
Animal development is commonly explained in terms of hierarchical genetic cascades that begin with an initial asymmetry based on either maternal or external cues. In Drosophila, for instance, complex cellular interactions during oogenesis eventually lead to the local deposition of maternal messages in the oocyte that determine the body axes (1, 2). In the Xenopus embryo, a pre-established animal-vegetal asymmetry in the egg and sperm entry point determine the location of the anterior-posterior and the dorsoventral axes by locally initiating genetic cascades that lead to the formation of the future organizer (3, 4). Less clear is how an organizer is set up in tissue without inherent asymmetry or external cues.
Only a few animal systems are amenable to the experimental analysis of self-organization during development (5–7). The simple metazoan Hydra is particularly useful in this context, because its body plan and any positional information can be completely destroyed and re-established in dissociation-reaggregation studies. Hydra consists of a single axis with a head and foot at either end of a tubular body column. The axial pattern of the animal is maintained by a gradient of head formation competence, commonly referred to as the head activation gradient (8), or source density gradient (9). The term gradient of head formation competence, or more simply gradient of head competence, is in line with current terminology and will be used in this work. (The term head activation will be used for the actual process of head formation.) This head competence gradient reflects the ability of tissue of the body column to form a head either on bisection leading to head regeneration at the apical end of the lower half, or on transplantation of a piece of the body column to the body column of a second animal. The gradient is maximal near the head decreasing down the body column and is a relatively stable property of the body column (8). Head formation in the body column is prevented by a head inhibition gradient, produced in the head and transmitted to the body column (10).
Tissue in which the head competence gradient has been destroyed can be generated by dissociating Hydra into a suspension of single cells and subsequently centrifuging them into a pellet, or aggregate. In these aggregates, new heads appear after 2–3 days and proceed to organize surrounding tissue into complete polyps (11–16). Previous work has shown that development of new heads and feet in aggregates occurs by true de novo pattern formation starting from nearly homogeneous conditions (12, 13). Because cells with different levels of head competence are randomly distributed throughout the aggregate, it is commonly assumed that head formation will occur where, by chance, cells with higher levels of head competence happen to be near one another. In such centers, termed activation centers, head activation is initiated, and they act as classical organizers that are able to recruit and instruct surrounding cells to participate in formation of the head as well as the whole axis. Here we show that de novo formation of activation centers minimally requires a cluster of 5–15 epithelial cells in which head activation is taking place. Molecular analysis shows that the conserved patterning genes HyBra1 and HyWnt are expressed in locally restricted areas corresponding to such clusters.
Materials and Methods
Animal Culture.
Polyps of the Basel strain of Hydra vulgaris were used for all experiments. Animals were maintained as described (12) in mass cultures, fed daily, and starved for 24 h before use in experiments.
Formation of Aggregates Containing Clusters of Vitally Labeled Cells.
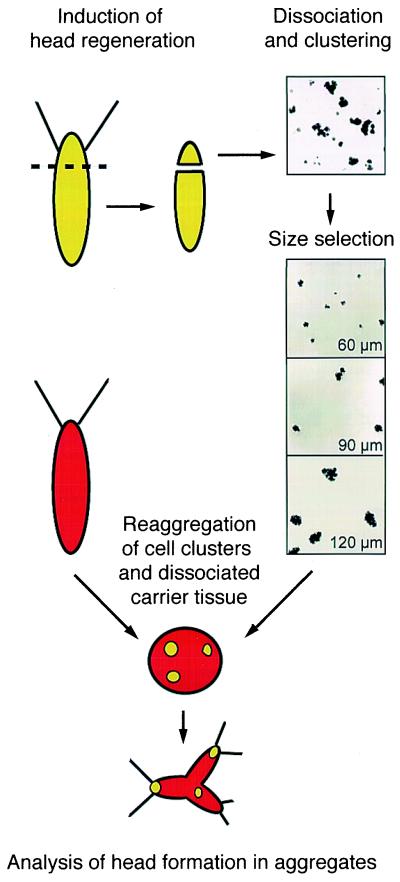
Animals were vitally labeled with FITC- or rhodamine-labeled beads (1-μm FITC-labeled or polychromatic Fluoresbrite plain microspheres; Polysciences) as described (12). About 50% of the epithelial cells were labeled with this procedure. To obtain cells with an elevated level of head activation, 300 labeled animals were decapitated and allowed to regenerate for 12 h. Regenerating tips, which are known to be undergoing head activation (8), were isolated and dissociated into a cell suspension by using dissociation medium as described (11). Clusters of cells formed by swirling the cell suspension (4 × 106 cells/ml) on a rotary shaker for 30 min at 10°C. The resulting suspension of cell clusters was size-fractionated on a 5% Percoll (Amersham Pharmacia) per dissociation medium column. The number of cells per cluster in each fraction was determined with a compound microscope. Cluster sizes of 60 ± 12 μm (10–25 cells), 90 ± 20 μm (40–60 cells), and 120 ± 31 μm (70–180 cells) were chosen for analysis. Cell clusters consisted primarily of endodermal and ectodermal epithelial cells with occasionally a few interstitial cells adhering to the periphery. To examine the possibility that dissociated cells were undergoing cell death, cell suspensions were stained with 4′,6-diamidino-2-phenylindole dihydrochloride and analyzed for signs of chromatin fragmentation (17). Within the time frame of the experiment, single cells, which make up 80–90% of the cell suspension used for aggregation, showed completely normal nuclear morphology and no indication of apoptosis.
To form aggregates containing clusters of labeled cells, the heads and feet of 300 budless polyps were removed, and the tissue was dissociated into cells. Then, statistically 1–2 labeled clusters of a specific size were added to an aliquot of the cell suspension and aggregates were formed. In addition to the labeled clusters, these aggregates contained 4,300 ± 500 unlabeled epithelial cells. During the development of the aggregates, the fraction of clusters found within developing heads was determined.
In Situ Hybridization.
Aggregates containing 8,000–10,000 epithelial cells were formed and allowed to develop. In situ hybridization analysis was performed as described (18) with slight modifications. RNA probes were prepared by amplifying directly from plasmids by PCR. They were used at a concentration of about 0.05 ng/μl.
Statistical Analysis.
Statistical significance was calculated by using the log likelihood ratio test (G test) based on calculated probabilities of binomial distributions of observed frequencies (19). The null hypothesis was rejected at the 95% confidence limit. Curve fitting was done by using igor pro 3.3 software (WaveMetrics, Lake Oswego, OR).
Results and Discussion
A Cluster of a Small Number of Cells from Head-Regenerating Tips Acts as an Organizer.
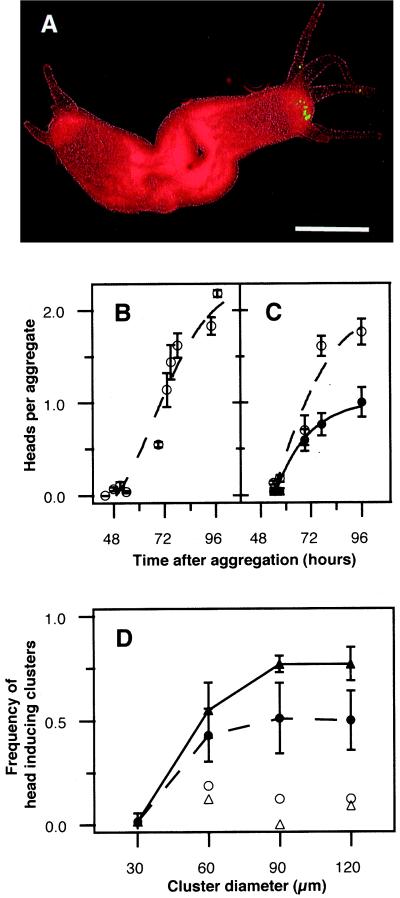
A crucial parameter of self-organization is the minimum number of cells that are necessary for the formation of a head activation center, or organizer. One approach to this question is to add clusters of different numbers of labeled cells undergoing head activation to an aggregate, and determine the minimum cluster size that is capable of inducing a head (Fig. 1) (see Materials and Methods). Both control aggregates without clusters and aggregates with clusters formed a total of two heads (Fig. 2 B and C). When labeled cell clusters were added to the carrier cell suspension, about half were found in a developing head (Fig. 2 A and C). This was true for all three cluster sizes (Fig. 2D). The effect was even more pronounced when the carrier tissue consisted of the lower four-fifths of the body column. In this case, two-thirds of the 90- and 120-μm clusters were in developing heads (Fig. 2D). Because the clusters contained only 0.2–4% of the cells in an aggregate, the high frequency of the appearance of the clusters in developing heads was not random. Instead, the results suggest that the cell clusters are involved in head induction.
Figure 1.
Experimental scheme of the self-organization assay in Hydra reaggregates; 12-h regenerating tips of vitally labeled polyps (green) were dissociated into single cells, and clusters were formed on a rotary cell shaker. Cell clusters were size-fractionated and clusters of each size were mixed with dissociated single-cell suspension from unlabeled carrier body columns (red) at statistically one to two clusters per aggregate. Head formation was followed individually in living aggregates over the next 96 h. For details, see Materials and Methods.
Figure 2.
Head induction by cell clusters. (A) A two-headed aggregate (96 h) with a head containing a green-labeled 60-μm cluster. (Bar = 200 μm.) (B) Time course of head formation in aggregates without introduced cell clusters. (C) Time course of the formation of heads containing 60-μm cell clusters (●) and total heads (○). Each data point is the mean ± SEM of 20–40 aggregates of three experiments. (D) Efficiency of cell clusters to induce head formation. Head formation frequency of single cells (30 μm) and different cell cluster sizes were scored 80 h after aggregation in carrier tissue derived from whole polyps (●; hatched line) and polyps lacking the upper fifth (▴; solid line) (P < 0.001, n = 28–53; means (SEM; three experiments). Control clusters derived from the corresponding carrier tissue are indicated by ○ and ▵.
In contrast, when 5–15 labeled single cells (each ≈ 30 μm in diameter) derived from regenerating tips were added to the cell suspension, virtually none were later found in developing heads (Fig. 2D). Similarly, when clusters were formed from cells of the body column that have a lower level of head competence, they were localized in developing heads at a low level (2–10%; Fig. 2D, open symbols), reflecting a statistical chance of being incorporated into a new head. A straightforward interpretation of these results is that clusters containing as few as 5–15 cells undergoing head activation, which corresponds to a cluster of 40–60 μm in diameter, have the capacity to act as a head-organizing center in an aggregate. The fact that a minimal number of cells undergoing head activation is necessary and sufficient to act as a head organizer suggests that a community effect (20) between these cells might be involved in the creation of an organizer.
Models based on reaction-diffusion mechanisms provide an explanation for the self-organization of an organizer (9, 21–25). These models assume a short-ranging activator and a long-ranging inhibitor. Because the production of the inhibitor is stimulated by the autocatalytic activator, they both have their maximum in the same location. In a field of virtually identical cells, random fluctuations in the activator concentrations are amplified by the autocatalysis of the activator leading to the generation of a stable pattern of local activation and lateral inhibition that functions as a prepattern for axial differentiation. The small cell clusters that were introduced in the experiment give a measure of the minimal requirements these random fluctuations have to meet to act as new organizers.
Besides the minimal organizer size, the range of the activator, which is the mean distance a molecule can travel between its production and disappearance (26), is of crucial importance for self-organization models. The range of the activator can be tentatively deduced as follows. The model predicts that fluctuations (i.e., clusters) smaller than the activator range will not be amplified because they lose activator too rapidly to the environment by diffusion. In contrast, cluster sizes that are similar to the activator range will amplify quite well, whereas clusters larger than the activator range will not have any further advantage in inducing heads. Thus, the rate of activation increase of an activation center, or cluster, is expected to reach a maximum once the cluster size is larger than the range of activation. This rate is crucial for the decision whether or not an activation center will be amplified or will decay. Accordingly, the activation range is probably not much larger than the size of a cluster (more precisely its radius) capable of inducing the formation of a head, which would be about 45 μm for a 90-μm cluster (about 2–3 epithelial cell diameters).
Clusters of Cells in Which Head Activation Occurs Inhibit the Formation of Heads Within Their Vicinity.
The second central component of a reaction-diffusion mechanism is the inhibitor that is produced by the activation center and transmitted to the surrounding tissue to prevent the initiation of another activation center. To obtain direct evidence concerning the presence and dimension of an inhibitory field, the following experiment was performed. A 120-μm cell cluster labeled red and a 60-μm cell cluster labeled green, both derived from regenerating tissue undergoing head activation, were added to carrier tissue and allowed to develop as usual. The larger clusters were found significantly more often in heads than were the smaller clusters (90% vs. 50%; Fig. 3 A and B; P < 0.01). This suggests that the larger cluster exerted an inhibitory influence on the smaller cluster. The failure or success of a 60-μm cluster to form a head was tightly correlated with its distance from the nearest head. About 50% of the 60-μm clusters were not involved in head formation at a distance of 600 μm, whereas essentially all were in heads at 1,000 μm (Fig. 3C). This indicates an effective range for the inhibition of about 800–900 μm.
Figure 3.
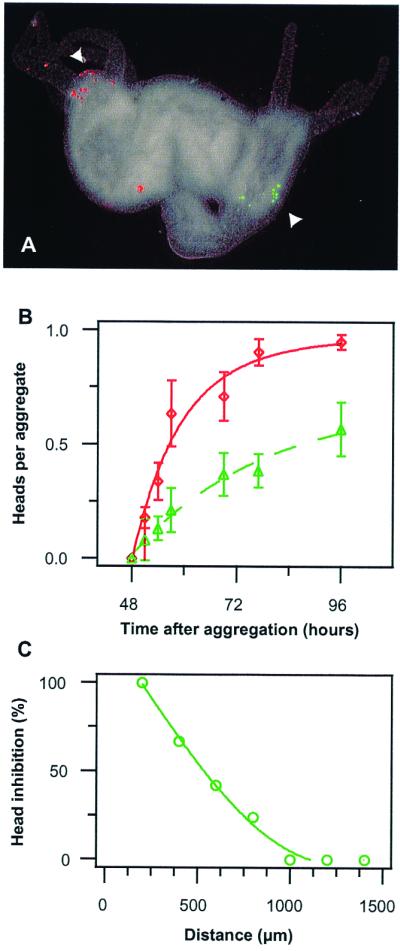
Effect of head inhibition in aggregates. (A) Aggregate (96 h) with a 120-μm cell cluster (red) that had induced a head (left arrow) and a 60-μm cell cluster (green) that did not induce a head (right arrow). (Bar = 200 μm.) (B) Time course of head induction by the 120-μm cell cluster (red circles) and the 60-μm cell cluster (green triangles) when competing in a single aggregate (means ± SEM), P < 0.001, n = 42–53, 3–5 experiments). (C) Correlation of head formation frequency of 60-μm cell clusters with their distance from the nearest head. The fate of cell clusters was determined at 96 h and the percentage of inhibited cell clusters for a given distance to the next head was calculated (n = 77).
A characteristic of a reaction-diffusion mechanism is that the range of inhibition is much greater than that of activation. The results obtained here are consistent with that condition. The range of activation, as deduced from the size of cell clusters able to induce head formation, is about 45 μm. In comparison, the range of inhibition (800–900 μm) is easily 20 times longer. These results are consistent with calculations based on a reaction-diffusion mechanism. In their pioneering paper, Gierer and Meinhardt (22) assume a 15-fold difference in diffusion constants between activator and inhibitor. Later, MacWilliams (8), to account for a variety of Hydra transplantation phenomena, assumed an even greater difference leading to a very small activated and large inhibited area in a proportion-regulating version of the Gierer-Meinhardt model.
In comparison, intact polyps can reach a size of about 2,000–4,000 μm before they start forming a second head during bud formation at a distance of about 2,000 μm from the adult head. Whereas this is beyond the inhibition range as determined in our aggregate assay, it is explained by the shallow gradient of the head competence, which reaches quite low levels at this distance from the head. At very low levels of head competence, the autocatalysis is less efficient and head activation less likely to occur, even at lowest levels of inhibition. In fact, Meinhardt (9) showed by modeling that the generation and maintenance of the graded competence is the critical factor for maintaining a single organizing region even when the length of the animal appears to exceed the range of the inhibition. Consistent with this view is that treatment with diacylglycerol (27), which raises the head competence gradient, results in the formation of ectopic heads along the body column.
Production of Head Inhibition Is Not Tightly Coupled with an Increase in Head Activation.
If the increase in lateral inhibition were tightly linked to the increase in activation, the spacing should be equal and more or less regular under all conditions, i.e., irrespective of the average level of head competence in the aggregate. We tested this prediction by analyzing aggregates made from tissue of either the apical halves (with a high level of head competence) or the basal halves (with a low level of head competence) of the body column. Several parameters indicated that the prediction was not met. Aggregates made from apical tissue formed more heads than did aggregates derived from basal tissue despite the fact that basal aggregates were 1.5 times larger on average (Table 1). This is also reflected in the average area per head, which is >4 times larger for heads in basal aggregates compared with those in apical aggregates. Were the increase in lateral inhibition closely coordinated with the increase in head activation, one would expect a similar average area per head.
Table 1.
Spacing of head formation in aggregates from apical and basal tissue
| Apical tissue | Basal tissue | |
|---|---|---|
| Heads per aggregate | 2.9 ± 1.5 | 1.1 ± 0.7 |
| Aggregate dimension, μm | ||
| Length | 1,000 ± 269 | 1,285 ± 305 |
| Width | 700 ± 149 | 898 ± 142 |
| Average aggregate area, μm2 | 19.1 × 105 | 31.3 × 105 |
| Minimal head distance, μm | 511 ± 248 | 1,237 ± 142 |
| Area per head, μm2 | 6.6 | 28.4 |
| Variance index | 0.49 | 0.12 |
The distance between each pair of heads in 15 aggregates of each type was measured 96 h after aggregate formation. The average area of an aggregate was calculated assuming it was an ellipsoid with the two axes corresponding is the average length and width of the aggregates. The variance index is SD mean value of the minimal distance between two heads. Error is the SD.
More significant is the highly irregular spacing of heads formed in apical tissue. Were the increase in lateral inhibition tightly linked to the increase in activation, one would expect the spacing between heads to be quite regular. Instead, the minimal distance in basal aggregates is twice as large as that in apical aggregates (Table 1). Perhaps the clearest measure of this greater irregularity of spacing of heads in apical tissue aggregates is the variance index (SD per mean value). Were heads spaced with perfect regularity, this index would have a value of zero. Instead, the variance index is four times higher in the apical aggregates than in basal tissue aggregates, indicating a much higher degree of irregularity in apical aggregates (Table 1).
These results suggest that the increase in inhibition is not tightly linked to the increase in activation, leading to a less rigid spacing in tissue with a high level of head competence. This is of particular relevance for the flexibility and robustness of the system, because it allows pattern formation to occur in a broad range of initial conditions. A similar conclusion concerning the increase in inhibition was drawn by MacWilliams (8) on the basis of transplantation experiments.
Two Hypostome Markers Indicating Cluster Behavior Reflect Organizer Formation Accurately.
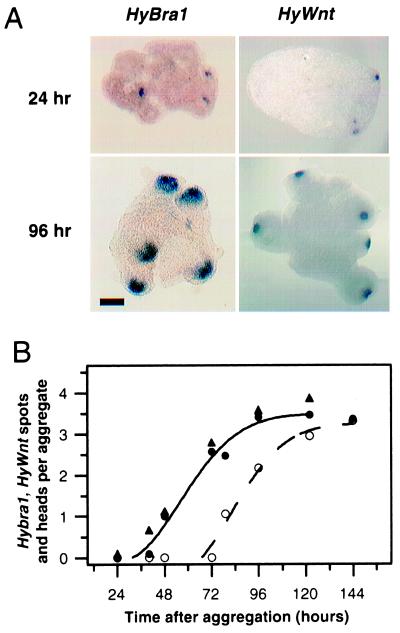
Finally, we wished to gain information about early molecular patterning events in the generation of self-organizing centers. To this end, we examined the expression pattern of two early head genes, HyBra1, a Hydra homologue of the T-box gene Brachyury (28), and HyWnt, a homologue of Wnt/Wg genes encoding signaling molecules (29). In intact polyps, these two genes have overlapping expression patterns in the head. Both are expressed very early during head regeneration, as well as during bud formation, the asexual form of reproduction of Hydra. In developing aggregates, the two layers, ectoderm and endoderm, have formed by 24 h (12). At this time, HyBra1 and HyWnt are first expressed almost simultaneously in small spots comprising only a few endodermal cells (Fig. 4 A and B). By 96 h, the HyBra1 and HyWnt spots have enlarged to their final size in future hypostomes (Fig. 4A). Interestingly, the size of the HyWnt spots are 50–60 μm, which corresponds to the minimal cluster size that acts as an organizer (Fig. 2D), whereas the HyBra1 spots (150–200 μm) are similar in size to the large clusters that can induce a head. Quantitative analysis shows that all spots eventually develop into heads about 2 days later (Fig. 4B).
Figure 4.
Early head patterning events visualized by the head markers HyBra1 and HyWnt. (A) In situ hybridization reveals early expression of the two hypostome markers in small spots (24 h), which enlarge during later stages (96 h). (Bar = 200 μm.) (B) Temporal analysis of head formation. HyBra1 (●) and HyWnt spots (▴) appear simultaneously and precede formation of morphological head structures (○) by about 2 days. All spots eventually develop into heads. Means of 25–82 aggregates from 2–5 experiments.
Subsequent to the early expression of these point-source genes, several other genes that are initially expressed uniformly throughout the aggregate become restricted to domains where new heads are being formed. Examples for this type of domain-restriction behavior are HyTCF and Hyβ-catenin, two other genes in the Wnt pathway (29), and the homeobox genes Cnox3 and msx (H.R.B., unpublished results). Thus, the results described above involving both the experiments with clusters of cells as well as those using molecular markers support the idea that the definition of point sources precedes the establishment of domains in the initially symmetrical environment of an aggregate.
Conclusions
We have shown that very small clusters of epithelial cells expressing the conserved HyBra1 and HyWnt genes act as head organizers during de novo pattern formation in Hydra aggregates. Members of the T-box gene family and Wnt pathway play crucial roles in the patterning of all higher animals (30, 31). The expression of these genes in the self-organization of head-organizing centers in aggregates of Hydra, a representative of one of the oldest metazoan phyla, strongly indicates the antiquity of these patterning systems (29, 32, 33). This pattern-forming system is able to generate complete structures (whole organisms) starting from a broad range of initial conditions. We therefore propose that during early metazoan evolution, an extremely robust and flexible self-organization system involving these molecular interactions was selected for, which became conserved during the evolution of higher animals.
Acknowledgments
We thank Patrick Lemaire for critical comments on the manuscript. This work was supported by a postdoctoral European Molecular Biology Organization long-term fellowship and Deutsche Forschungsgemeinschaft Grants Te 311/1–2 to U.T. and SFB 474-B1 to B.H. and T.W.H, and National Science Foundation Grants IBN-9723660 and IBN-9904757 to H.R.B.
References
- 1.Ray R P, Schüpbach T. Genes Dev. 1997;10:1711–1723. doi: 10.1101/gad.10.14.1711. [DOI] [PubMed] [Google Scholar]
- 2.St. Johnston D, Nüsslein-Volhard C. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 3.Moon R T, Kimelman D. BioEssays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 4.Harland R, Gerhart J. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 5.Psychoyos D, Stern C D. Development (Cambridge, UK) 1996;122:3263–3273. doi: 10.1242/dev.122.10.3263. [DOI] [PubMed] [Google Scholar]
- 6.Yuan S, Schoenwolf G C. Development (Cambridge, UK) 1998;125:201–213. doi: 10.1242/dev.125.2.201. [DOI] [PubMed] [Google Scholar]
- 7.Bernacki S H, McClay D R. J Exp Zool. 1989;251:203–216. doi: 10.1002/jez.1402510208. [DOI] [PubMed] [Google Scholar]
- 8.MacWilliams H K. Dev Biol. 1983;96:239–257. doi: 10.1016/0012-1606(83)90325-1. [DOI] [PubMed] [Google Scholar]
- 9.Meinhardt H. Dev Biol. 1993;157:321–333. doi: 10.1006/dbio.1993.1138. [DOI] [PubMed] [Google Scholar]
- 10.MacWilliams H K. Dev Biol. 1983;96:217–238. doi: 10.1016/0012-1606(83)90324-x. [DOI] [PubMed] [Google Scholar]
- 11.Gierer A, Berking S, Bode H R, David C N, Flick K M, Hansmann G, Schaller H, Trenkner E. Nat New Biol. 1972;239:98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- 12.Technau U, Holstein T W. Dev Biol. 1992;151:117–127. doi: 10.1016/0012-1606(92)90219-7. [DOI] [PubMed] [Google Scholar]
- 13.Sato M, Tashiro H, Oikawa A, Sawada Y. Dev Biol. 1992;151:111–116. doi: 10.1016/0012-1606(92)90218-6. [DOI] [PubMed] [Google Scholar]
- 14.Lee P-C, Javois L. Dev Biol. 1993;157:10–18. doi: 10.1006/dbio.1993.1107. [DOI] [PubMed] [Google Scholar]
- 15.Sato M, Bode H R, Sawada Y. Dev Biol. 1990;141:412–420. doi: 10.1016/0012-1606(90)90395-y. [DOI] [PubMed] [Google Scholar]
- 16.Sato M, Sawada Y. Dev Biol. 1989;133:119–127. doi: 10.1016/0012-1606(89)90303-5. [DOI] [PubMed] [Google Scholar]
- 17.Cikala M, Wilm B, Hobmayer E, Böttger A, David C N. Curr Biol. 1999;9:959–962. doi: 10.1016/s0960-9822(99)80423-0. [DOI] [PubMed] [Google Scholar]
- 18.Grens A, Mason E, Marsh J L, Bode H R. Development (Cambridge, UK) 1995;121:4027–4035. doi: 10.1242/dev.121.12.4027. [DOI] [PubMed] [Google Scholar]
- 19.Sokal R R, Rohlf F J. Biometry. 3rd Ed. New York: Freeman; 1995. pp. 1–685. [Google Scholar]
- 20.Gurdon J, Lemaire P, Kato K. Cell. 1993;75:831–834. doi: 10.1016/0092-8674(93)90526-v. [DOI] [PubMed] [Google Scholar]
- 21.Turing A. Philos Trans R Soc London B. 1952;237:37–72. [Google Scholar]
- 22.Gierer A, Meinhardt H. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 23.Meinhardt H. Models of Biological Pattern Formation. London: Academic; 1982. [Google Scholar]
- 24.Kauffman S. The Origin of Order. London: Oxford Univ. Press; 1993. [Google Scholar]
- 25.Harrison L G. Kinetic Theory of Living Pattern. New York: Cambridge Univ. Press; 1993. [Google Scholar]
- 26.Meinhardt H, Gierer A. BioEssays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 27.Müller W A. Development (Cambridge, UK.) 1989;105:309–316. [Google Scholar]
- 28.Technau U, Bode H R. Development (Cambridge, UK) 1999;126:999–1010. doi: 10.1242/dev.126.5.999. [DOI] [PubMed] [Google Scholar]
- 29.Hobmayer B, Rentzsch F, Kuhn K, Happel C M, Cramer von Laue C, Snyder P, Rothbächer U, Holstein T W. Nature (London) 2000;407:186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 30.Smith J. Trends Genet. 1999;15:154–159. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- 31.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 32.Bode H, Martinez D, Shenk M A, Smith K, Steele R, Technau U. Biol Bull (Woods Hole, MA) 1999;195:408–410. doi: 10.2307/1542982. [DOI] [PubMed] [Google Scholar]
- 33.Galliot B, Miller D. Trends Genet. 2000;16:1–5. doi: 10.1016/s0168-9525(99)01888-0. [DOI] [PubMed] [Google Scholar]