Abstract
The origin of the eukaryotic cell remains an unsolved question. Numerous experimental and phylogenetic observations support the symbiotic origin of the modern eukaryotic cell, with its nucleus and (typically) mitochondria. Incorporation of mitochondria has been proposed to precede development of the nucleus, but it is still unclear whether mitochondria were initially part of basal eukaryotes. Data on alanyl-tRNA synthetase from an early eukaryote and other sources are presented and analyzed here. These data are consistent with the notion that mitochondrial genesis did not significantly precede nucleus formation. Moreover, the data raise the possibility that diplomonads are primary amitochondriates that radiated from the eukaryotic lineage before mitochondria became fully integrated as a cellular organelle.
Functional and phylogenetic studies of mitochondrial genomes clearly demonstrate an eubacterial origin for mitochondria (1–8). However, the scarceness of reliable molecular markers for the initial events that gave rise to the eukaryotic lineages has prevented evolutionary biologists from obtaining detailed information about early events in eukaryote cell formation. The most apparent consequence of the integration of mitochondria within the eukaryotic cell is the gross reduction in size and gene content of the eubacterial symbiont's genome (9). During this process, redundancy in enzyme families was eliminated, presumably to avoid duplication and metabolic incongruencies. This process of genome integration is not understood, but must have required a gradual genetic transfer from the premitochondrial genome to the nuclear genome of the preeukaryotic cell. During this flow of genetic information, the bacterial endosymbiont evolved into a cellular organelle.
This process offered the potential for the generation of mosaic-like organisms, defined here as species that underwent the initial stages of the genetic exchange process, but evolved away from the main eukaryote line of descent, without fully incorporating mitochondria. These organisms would contain genomic evidence (in the form of genes of mitochondrial origin) of the initial genetic transfers that occurred between the premitochondrial endosymbiont and the nuclear genome. However, other genes in these species (which were later replaced in other eukaryotes by their mitochondrial equivalents) would have remained nuclear in nature.
Aminoacyl-tRNA synthetases (aaRS), enzymes responsible for the aminoacylation of tRNAs, represent a case in point for this process of genomic integration. As essential components of the translational apparatus, the genes coding for aaRS in the nucleus of the first eukaryotic cells were of archaeal origin (10, 11). The incorporation of the mitochondrial genome into the eukaryotic cell (with its own set of genes coding for aaRS) resulted in the integration of these two gene families in the nuclear genome. Analysis of sequences of eukaryotic aaRS (including nuclear-encoded mitochondrial enzymes) shows that the mitochondrial aaRS genes were either transferred to the nucleus (where they either coexist with their nuclear cytoplasmic counterparts or have replaced them) or were lost and functionally replaced by their nuclear equivalents (10, 11).
These considerations suggest that a close examination of the evolutionary history of a specific tRNA synthetase could lead to insights into the establishment of mitochondria in the eukaryote cell. In particular, we were interested in the possibility of an early amitochondriate eukaryote housing an archaeal-like rather than a mitochondrial-like gene for a particular synthetase. To that end, we analyzed the phylogenetic relationships of the cytoplasmic and mitochondrial alanyl-tRNA synthetases (AlaRS) to determine the evolutionary pattern of distribution of this enzyme in mitochondriate and in amitochondriate eukaryotes.
Materials and Methods
Cloning and Sequencing alaS Genes.
Giardia lamblia genomic DNA was a gift from genomic DNA provided by F. Gillin (University of California, San Diego). Sequences corresponding to the termini of the gene encoding AlaRS were identified in contigs generated by the Giardia sequencing project. Primers corresponding to these sequences were used to amplify the gene from genomic DNA. The 2907-bp PCR product was subcloned into pCR-2.1-Topo (Invitrogen), and the insert was completely sequenced.
blast searches against expressed sequence tag (EST) databases indicated that Drosophila melonogaster cDNA clones LD11251 and LD07142 encoded AlaRS. The clones were obtained from the Berkeley Drosophila Genome Project. Primers based on the λ-ZAP vector were used to amplify the cDNA inserts, and the corresponding PCR products were subcloned into pCR-2.1-Topo and completely sequenced. Sequence homology, phylogenetic analysis, and the presence of a 5′ leader sequence in clone LD11251, which is thought to encode a mitochondrial targeting sequence, indicate that LD11251 encodes the mitochondrial synthetase and LD07142 the cytoplasmic enzyme. Dictyostelium discoideum cDNA clone C25775 was also identified by using blast searches as sequence coding for AlaRS. The clone was obtained from H. Urushihara (Tsukuba University, Tsukuba, Japan), and the insert was completely sequenced.
Phylogenetic Analysis.
Publicly available databases, including partial genomic sequences, were searched for homologous ORFs to AlaRS from Escherichia coli, Archaeoglobus fulgidus, and Saccharomyces cerevisiae by using the programs blastp, tblastn, and psi-blast (12). Amino acid sequences were first aligned by using the program clustalw with the default parameters (13) and then manually edited.
All indels (insertions and deletions) were omitted from the multiple sequence alignment before phylogenetic analysis. Maximum parsimony analysis was done by using the software package paup* 4.0 (23). Given the large size of the two datasets, it was not possible to exhaustively search for the total number of minimal length trees. Instead, the numbers and lengths of minimal trees were estimated from 100 replicate random heuristic searches, whereas confidence limits of branch points were estimated by 1,000 bootstrap replications.
The neighbor-joining phylogeny was based on pairwise distances between amino acid sequences by using the programs neighbor and protdist of the phylip 3.57c package (22). The “Dayhoff” program option was invoked in the latter program, which estimates the expected amino acid replacements per position by using a replacement model based on the Dayhoff 120 matrix. The programs seqboot and consense were used to estimate the confidence limits of branching points from 1,000 bootstrap replications.
The maximum likelihood phylogeny was done by using the program puzzle 4 (14). The invoked options were 1,000 puzzling steps, the JTT substitution matrix, and the γ distribution model with eight rate categories for estimation of rate heterogeneity, and α parameter estimation from the dataset.
The significance of alternative, but less optimal, tree topologies was tested by using the methods of Kishino-Hasegawa and Templeton as implemented in the software packages puzzle and phylip (protpars program), respectively. Topologies tested against included one where Giardia lamblia was the basal lineage in a clade of all eukaryotes, the canonical view of the evolutionary status of G. lamblia (15).
Results
We completely sequenced strategically chosen genes for AlaRS (alaS) in the amitochondriate G. lamblia (genomic DNA provided by F. Gillin), the mitochondriate slime mold D. discoideum (clone from H. Urushihara), and the nuclear-encoded mitochondrial alaS gene of D. melanogaster (clone from the Berkeley Drosophila Genome Project).
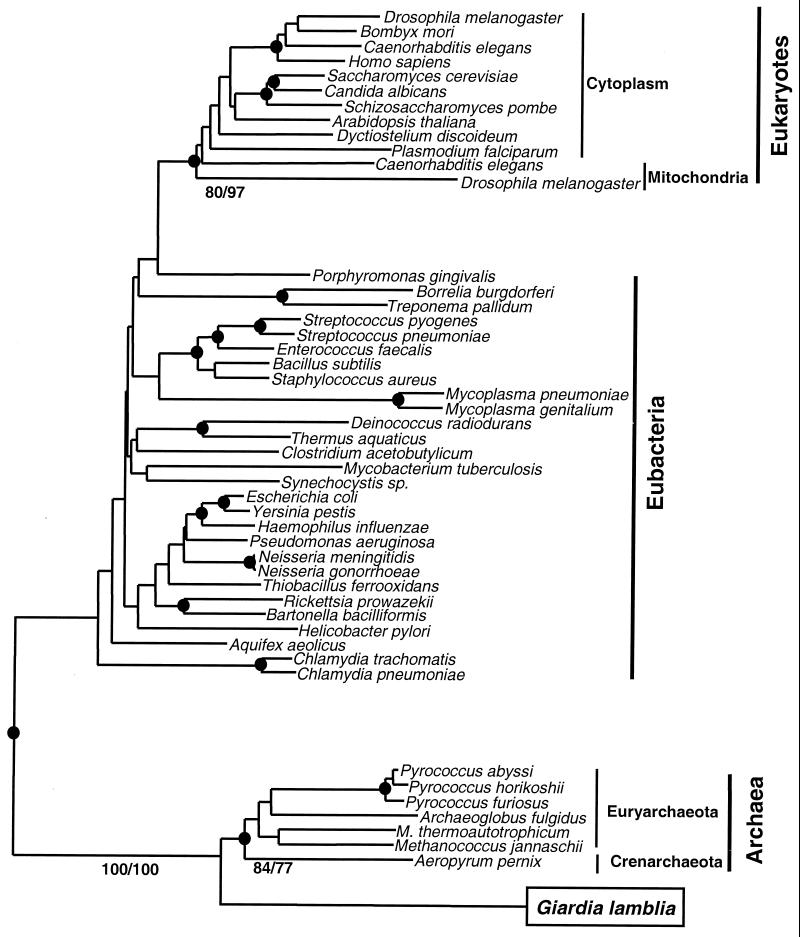
We found that all sequences of the nuclear-encoded cytoplasmic AlaRSs from mitochodriate eukaryotes were rooted by mitochondrial AlaRSs. Furthermore, both mitochondrial and cytoplasmic AlaRSs are nested within the eubacterial AlaRSs. These results indicate that all AlaRSs of extant mitochondriate eukaryotes are of mitochondrial origin, probably because of a replacement of the original cytoplasmic alaS by its mitochondrial equivalent. The clustering of the eukaryotic AlaRS sequences within the bacterial branch is in agreement with the endosymbiotic theory for the origin of mitochondria (Fig. 1).
Figure 1.
Phylogenetic analysis of AlaRS sequences. Neighbor-joining (NJ) tree constructed by using the programs neighbor and protdist (24), where the number of amino acid substitutions per site in pairwise comparisons of aligned sequences (415 amino acids in length) were estimated by using the Dayhoff 120 matrix (22). The position of Giardia AlaRS is boxed. A similar tree topology was determined in 100 replicate random heuristic searches by using the maximum parsimony (MP) method (24) (one minimal length tree, 4821 steps long). ●, Those nodes supported in 70% or greater of 1,000 random bootstrap replicates of both MP and NJ trees. Bootstrap values for critical nodes are shown as % of 1,000 replicates in MP/NJ methods. The node leading to G. lamblia and archaeal species was consistently supported in 100% of replications. Furthermore, the shown tree topology was significantly better statistically [Kishino-Hasegawa and Templeton tests (25); P < 0.05] than all other alternative topologies of the major groups, including one where eukaryotes and G. lamblia were clustered together.
Surprisingly, our analyses revealed that the AlaRS from the eukaryote G. lamblia constitutes an exception, being more closely related to archaeal AlaRS than to either eubacterial or other eukaryotic AlaRSs (Fig. 1). The complete agreement among different phylogenetic methods (protein maximum likelihood, maximum parsimony, and neighbor-joining distance methods gave identical topologies for the eukaryotic AlaRS sequences), and the existence of clear synapomorphies (Fig. 2) support this finding. Thus, the alaS gene shows different patterns of acquisition and retention between an amitochondriate and mitochondriate eukaryotes. This unique evolutionary pattern sets the AlaRS-based phylogenies apart from those obtained for other aaRS (10, 11, 16).
Figure 2.
Synapomorphic insertion in AlaRSs. This insertion is located within the conserved catalytic domain of AlaRS (26), and unites mitochondrial, cytoplasmic (labeled E), and eubacterial AlaRSs (labeled B), but is missing in archaeal (labeled A) and Giardia AlaRSs. Archaeal and Giardia sequences are shadowed in purple, and the position of Giardia AlaRS is boxed. Multiple sequence alignments and phylogenies are available from the authors. The sequences determined here have been deposited in GenBank (accession nos. AF188719, AF188717, and AF188716).
Discussion
The basal position of the mitochondrial alaS genes within the eukaryotic branch clearly supports a replacement of a preexisting nuclear (or archaeal-like) alaS gene by its mitochondrial counterpart. With the current set of sequences, we can establish that this replacement at least preceded the divergence of the eukaryote phyla Rhizopoda (Dictyostelium) and Apicomplexa (Plasmodium) from the main eukaryote lineage. The discovery here that most eukaryote AlaRSs are of mitochondrial origin will require a reexamination of details and features of the sequence conservation of AlaRS, which previously was considered one of the most conserved of all aaRSs that could not be differentiated into groups (e.g., bacterial, archaeal, etc.) (17).
The process of extensive lateral gene transfers that resulted in the reduction of mitochondrial genome size must have required a large amount of time. However, it is likely that integration of nuclear and the proto-mitochondrial genes involved in translation (including alaS) took place at an even earlier stage of eukaryote evolution, because early sorting of metabolic duplications must have been a requirement for the functional fusion of the mitochondrial and the nuclear genomes.
Two potential scenarios can explain the atypical phylogenetic position of Giardia alaS. The first possibility is that Giardia alaS is the result of a late lateral gene transfer from archaea, which occurred after the hypothetical loss of mitochondria by Giardia. If so, then Giardia alaS would represent a novel and interesting example of archaea to eukaryote gene transfer. This possibility is unlikely for two reasons. First, it requires two rounds of gene transfer and replacements. Namely, the replacement of the nuclear alaS gene by the mitochondrial equivalent (as seen in the rest of eukaryote organisms); and a posterior replacement (only in diplomonads) of the mitochondrial alaS gene by an archaeal alaS. The second difficulty is that, in all of our phylogenetic trees, Giardia AlaRS is placed significantly before the deepest archaeal split that separates the kingdoms crenarchaeota and euryarcheota (Fig. 1). This observation suggests that the Giardia alaS gene is not the result of a late lateral gene transfer, such as that seen in the case of bacterial GluRS sequences (18).
The second scenario is that the Giardia genome did not experience the same extent of genetic transfer between mitochondria and nuclei that other eukaryotic organisms underwent. If this is the case, then diplomonads would have diverged away from the eukaryotic lineage before a full integration of the mitochondrial genome was accomplished, and its alaS gene would have remained archaeal in nature, because it would not have been replaced by a mitochondrial alaS gene.
This second explanation is simple, and consistent with a gradual course of endosymbiosis and integration of mitochondrial genes. In this context, Giardia would represent an intermediate stage of the process. (The full integration of mitochondria could then result in the nuclear substitution of the archaeal alaS gene by the mitochondrial alaS.) Moreover, this hypothesis allows for an interpretation of our results compatible with those obtained by studying the phylogenetic relationships of other Giardia genes. The analysis of Giardia valyl-tRNA synthetase and heat shock proteins, for instance, clearly indicates that lateral gene transfer occurred between the ancestral genomes of extant mitochondria and the ancestral nuclear genome of Giardia (19, 20). Because this process of transfer did not include the alaS gene, the data may be indicating an early radiation of diplomonads away from the main eukaryote line, after the first genetic transfers from the premitochondrial genome occurred, but before the completion of mitochondria integration.
Our results offer an evolutionary scenario for the eukaryote cell that needs to be contrasted with previous and future data (2, 5, 7, 8, 19–21). We propose that diplomonads radiated from the eukaryotic line of descent before the complete integration of the mitochondrial ancestor with its host. Indeed, although Giardia alaS has not been replaced by its mitochondrial equivalent, other genes in this species originate from ancestors of extant mitochondria (19, 22, 23). If the diplomonads represent an intermediate stage in mitochondrial genesis, and considering the nucleated nature of Giardia, then the appearance of mitochondria may not have preceded the genesis of modern nuclei. It is of great interest to investigate whether the evolutionary history of Giardia AlaRS reported here is also seen with data from other diplomonads and basal eukaryotes.
Acknowledgments
We thank Drs. F. Gillin (University of California, San Diego) and H. Urushihara (Tsukuba University, Tsukuba, Japan), and the Berkeley Drosophila Genome Project for providing the genetic material used in this work. We also thank Dr. Gillin and members of her laboratory for technical and scientific discussions. This work was supported by Grant GM23562 from the National Institutes of Health and by a fellowship from the National Foundation for Cancer Research.
Abbreviations
- aaRS
aminoacyl-tRNA synthetase
- AlaRS
alanyl-tRNA synthetase
Footnotes
Data deposition: The sequences presented in this paper have been deposited in the GenBank database (accession nos. AF188719, AF188717, and AF188716).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220388797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220388797
References
- 1.Cavalier-Smith T. Nature (London) 1987;326:332–333. doi: 10.1038/326332a0. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. Ann NY Acad Sci. 1987;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- 3.Cavalier-Smith T. Ann NY Acad Sci. 1987;503:55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- 4.Margulis L. BioSystems. 1993;31:121–125. doi: 10.1016/0303-2647(93)90039-f. [DOI] [PubMed] [Google Scholar]
- 5.Margulis L. Proc Natl Acad Sci USA. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woese C R. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin W, Muller M. Nature (London) 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 8.Vellai T, Takacs K, Vida G. J Mol Evol. 1998;46:499–507. doi: 10.1007/pl00006331. [DOI] [PubMed] [Google Scholar]
- 9.Lang B F, Burger G, O'Kelly C J, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Gray M W. Nature (London) 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 10.Brown J R, Doolittle W F. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf Y I, Aravind L, Grishin N V, Koonin E V. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 12.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strimmer K, von Haeseler A. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalier-Smith T. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woese C R, Olsen G J, Ibba M, Soll D. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiba K, Motegi H, Schimmel P. Trends Biochem Sci. 1997;22:453–457. doi: 10.1016/s0968-0004(97)01135-3. [DOI] [PubMed] [Google Scholar]
- 18.Lamour V, Quevillon S, Diriong S, N′Guyen V C, Lipinski M, Mirande M. Proc Natl Acad Sci USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto T, Sanchez L B, Shirakura T, Muller M, Hasegawa M. Proc Natl Acad Sci USA. 1998;95:6860–6865. doi: 10.1073/pnas.95.12.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta R S, Aitken K, Falah M, Singh B. Proc Natl Acad Sci USA. 1994;91:2895–2899. doi: 10.1073/pnas.91.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreira D, Lopez-Garcia P. J Mol Evol. 1998;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- 22.Roger A J, Svard S G, Tovar J, Clark C G, Smith M W, Gillin F D, Sogin M L. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horner D S, Hirt R P, Kilvington S, Lloyd D, Embley T M. Proc R Soc London Ser B. 1996;263:1053–1059. doi: 10.1098/rspb.1996.0155. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 25.Swofford D L. paup* Sunderland, MA: Sinauer; 1999. , Version 4.0. [Google Scholar]
- 26.Ribas de Pouplana L, Buechter D D, Davis M W, Schimmel P. Protein Sci. 1993;2:2259–2262. doi: 10.1002/pro.5560021225. [DOI] [PMC free article] [PubMed] [Google Scholar]