Abstract
The cell wall of bakers' yeast contains a family of glycosyl-phosphatidylinositol (GPI)-linked glycoproteins of domain structure similar to the adhesins of pathogenic fungi. In wild-type cells each of these proteins has a unique function in different developmental processes (mating, invasive growth, cell–cell adhesion, or filamentation). What unifies these developmental events is adhesion, either to an inert substrate or to a cell. Although they differ in their specificities, many of these proteins can substitute for each other when overexpressed. For example, Flo11p is required during vegetative growth for haploid invasion and diploid filamentation, whereas Fig2p is required for mating. When overexpressed, Flo11p and Fig2p can function in mating, invasion, filamentation, and flocculation. The ability of Flo11p to supply Fig2p function in mating depends on its intracellular localization to the mating projection, where Fig2p normally functions in the adhesion of mating cells. Our data show that even distant family members retain the ability to carry out disparate functions if localized and expressed appropriately.
A family of glycosyl-phosphatidylinositol (GPI)-linked cell surface glycoproteins termed the fungal adhesions are important for fungal pathogenesis because they permit pathogens such as Candida albicans to adhere to mammalian epithelial and endothelial cells (1, 2). These proteins have an N-terminal signal sequence, a central domain containing a highly repeated serine/threonine-rich sequence, and a C-terminal domain containing a GPI anchoring sequence. Expression of these proteins [e.g., Als1p from Candida albicans (3, 4) and Epa1p from Candida glabrata (5)] in Saccharomyces permits this organism, which does not normally adhere to mammalian cells, to adhere to them.
The Saccharomyces cerevisiae genome contains a family of cell wall proteins related to the adhesins of pathogenic fungi. One branch of this protein family, encoded by genes including FLO1, FLO5, FLO9, and FLO10, is called the flocculins (6) because these proteins promote cell–cell adhesion to form multicellular clumps that sediment out of solution (7–10). The FLO1, FLO5, FLO9, and FLO10 genes share considerable sequence homology.
A second group of Flo family members has a domain structure similar to that of the first, but with quite unrelated amino acid sequences. This second group includes three proteins, Flo11p, Fig2p, and Aga1p. Flo11p is required for diploid pseudohyphal formation and haploid invasive growth (11, 12). In haploid invasive growth cells adhere to the agar surface, so that they do not wash off (13). In diploid pseudohyphal growth cells adhere to each other after division and form long chains of filaments (14). Fig2p and Aga1p are induced during mating (15, 16). Aga1p, linked by disulfides to the soluble peptide, Aga2p (17), is required on the surface of MATa cells for them to adhere to the protein Sag1p on the surface of MATα cells (18). Aga1p, Aga2p, and Sag1p are required for mating in liquid but not solid medium.
These morphogenetic events—flocculation, filamentation, invasive growth, and mating—require very different signaling pathways and cellular structures. Flocculation often occurs upon depletion of sugar during late-exponential or stationary phases of growth (19–21), whereas filamentation requires starvation for nitrogen (14). Mating requires the cessation of growth and induction of mating-specific genes by mating pheromones (22–25). The cell biology of the four morphogenic events is also different. Flocculation and haploid invasion are isotropic; there is no evidence for regional localization of the adherent surface. By contrast, mating and pseudohyphal filamentation require polarized growth—mating cells produce projections that are the sites of cell adherence and fusion, whereas pseudohyphal growth requires cells to stick together at their ends.
In this report we show that the flocculin genes of yeast form an unusual network of functionally interrelated family members. These genes all have a similar overall domain structure, but they have considerable diversity in amino acid sequence. Given these differences, it is striking that one FLO gene can compensate for another in diverse morphogenic events: flocculation, mating, haploid invasion, and filamentation. What unifies these developmental events is adhesion, either to an inert substrate or to a cell.
Materials and Methods
Yeast Strains and Growth Conditions.
All yeast strains (Table 1 in supplementary material at www.pnas.org) used in this study are congenic to the Σ1278b genetic background (26, 27). Standard yeast culture medium was prepared as described (28). All yeast strains were grown at 30°C. Synthetic low-ammonia dextrose (SLAD) medium and synthetic low-ammonia raffinose (SLAR) medium used to induce pseudohyphae were prepared (SLAD = 2% glucose; SLAR + Gal = 2% raffinose + 0.2% galactose) as described (14). Pseudohyphal filamentous growth was determined by streaking strains on SLAD or SLAR + Gal plates and photographing them after 4–7 days of growth. Pseudohyphal filaments that invaded agar were observed after washing the plates with water. The invasive growth assays were carried out by washing the plates under the tap (13) after 5 days of growth on yeast extract/peptone/dextrose (YPD) or yeast extract/peptone Gal (YPGal).
Primers and Plasmid Construction.
All primers are listed in Table 2 in supplementary material at www.pnas.org.
pQF142.1 (plasmid for deletion of FLO11): A 2 μ plasmid, pQF146, containing the entire open reading frame (ORF) of FLO11 (from nucleotide 1471 upstream to nucleotide 1599 downstream) was isolated from an S. cerevisiae genomic library (C. Connelly and P. Hieter, personal communications). A BlpI/NruI fragment of pQF146 including the entire FLO11 ORF was replaced with a HIS3 gene fragment to yield pQF142.1.
pQF296.10 (plasmid for FLO11–3HA and FIG2–3HA constructs): S. cerevisiae URA3 gene was amplified by PCR with primers BG118 and BG119. The PCR product was digested with NotI/XbaI and cloned into pBluescript KSII(+) NotI/XbaI sites to form pQF292. The triple hemagglutinin epitope (HA) tags from B2385 (NotI fragment, Fink laboratory collection) and from B2500 (XbaI fragment, Fink laboratory collection) are cloned into NotI and XbaI sites of pQF292 sequentially to form pQF296.10. The reading frame of the HA tag is from NotI to XbaI.
Construction of Strains with GAL1 Promoter Inserted in Front of the Cell Wall Protein-Encoding Genes.
Primer pairs were used to amplify a plasmid (pFA6a-KanMX6-pGAL1) containing the GAL1 promoter and the kanamycin-resistance gene (29). PCR products were used to transform the appropriate yeast strains, and kanamycin-resistant transformants were checked by PCR to make sure the insertion occurred in the correct location. The primer pairs for different genes are as follows: FIG2, BG88 and BG89; FLO1, BG94 and BG95; FLO10, BG96 and BG97; and FLO11, BG156 and BG157.
Genetic Crosses, Transformation, and Mating.
For most of the experiments, standard methods for genetic crosses and transformation were used (30). To make the diploid strains WY454, WY449, WY452, WY450, WY451, WY415, and WY453, haploid MATa strains of the appropriate genotype were transformed to Ura+ (uracil independence) with YCP50-HO. Most of the Ura+ transformants were diploids as a result of mating type switching and mating within the clone. The transformants were streaked to a plate containing 5-fluoroorotic acid (5-FOA) (31) to get rid of the YCP50-HO plasmid. Quantitative mating assays were performed essentially as described (32). The mating efficiency was defined as the number of diploid colonies divided by the combined number of colonies formed by the haploid parent being tested and the number of diploids.
Gene Knockouts.
flo11∷HIS3 knockout was achieved by transformation of a wild-type MATa haploid with a XbaI/EcoRV-digested plasmid pQF142.1. Gene knockouts for the following genes were carried out by transformation with the appropriate PCR products. The primer pairs and the cognate templates are as follows: fig2∷URA3, BG84 and BG85, pRS316; fig2∷TRP1, BG84 and BG85, pRS314; aga1∷HIS3, BG68 and BG69, B3651; aga2∷HIS3, BG110 and BG111, B3651; sag1∷HIS3, BG72 and BG73, B3651; and flo10∷URA3, BG142 and BG143, pRS316.
Preparation of RNA and Northern Hybridization.
Total RNA was prepared by using hot acid phenol, and Northern blotting was performed as described (33). Overnight cultures of yeast strains in YPD were diluted 20-fold with YPD and grown to OD600 = 1.0. These cultures were centrifuged and RNA was prepared. A 10-μg sample of RNA was run on a gel, blotted, and hybridized with a 500-bp fragment of FLO10 (corresponding to the N-terminal ORF sequence) probe or a 500-bp fragment of FLO11 (corresponding to the N-terminal ORF sequence) probe.
Tagging Flo11p and Fig2p with 3HA.
A 3HA-URA3–3HA-encoding region in plasmid pQF296.10 was amplified by primers BG122 and BG123 (for tagging Flo11p) or primers BG120 and BG121 (for tagging Fig2p) The PCR products were used to transform strain 10560–2B. The correct transformants with 3HA-URA3–3HA inserted at the ORF region of FLO11 or FIG2 were streaked to 5-fluoroorotic acid plates (31) to select for isolates that have looped out the URA3 gene by homologous recombination.
Indirect Immunofluorescence.
Yeast cells were removed from the surface or from beneath the agar medium with toothpicks and stained as follows: Yeast strains WY423 (haploid) and WY453 (diploid) grown on synthetic complete medium or WY453 grown on SLAD were fixed in phosphate-buffered saline (PBS) containing formaldehyde (3.7%) for 1 h, washed with PBS, and washed again with PBS containing 2% bovine serum albumin. After incubation in PBS containing 2% albumin for 1 h, cells were precipitated and resuspended in PBS containing 2% albumin and mouse anti-HA antibody (1/1000) for 1 h. Cells were then washed with PBS containing 2% albumin three times and resuspended in PBS containing 2% albumin and Cy3-conjugated goat anti-mouse IgG antibody (1/1000) for 20 min. Cells were then washed three times with PBS containing 2% albumin.
To stain yeast cells that had been induced with pheromone, overnight cultures of strains WY423, WY427, and WY491 were diluted 50-fold in YPD, grown for 4 h, and treated with 5 μg/ml α factor for 1 h (WY491 was washed and transferred to YPGal before pheromone treatment). The cultures were treated with another addition of 5 μg/ml α factor and incubated for 1 more hour. The cultures were sonicated for 30 sec and fixed by addition of 1/10 vol of 37% formaldehyde for 2 h. Then cells were washed with PBS containing 2% albumin and stained as described previously.
Results
Overexpression of Cell Wall Proteins Causes Flocculation and Adherence to the Agar.
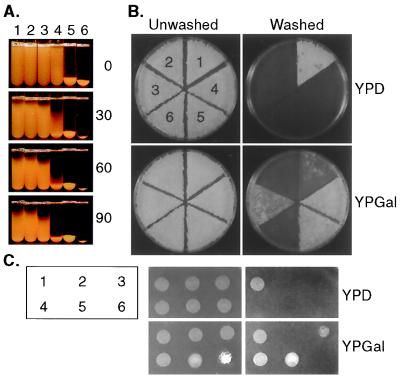
The enhanced expression of any one of a number of cell wall proteins results in aggregation of the cells because of cell–cell adhesion. This phenomenon is easily observed in liquid because cultures of the Σ1278b strain take hours to clarify, whereas flocculent strains begin to sediment much more rapidly. Isogenic Σ1278b strains overexpressing FIG2, FLO11, FLO10, or FLO1 were compared for the time it took for each culture to sediment. To make this comparison, the GAL1 promoter was inserted at the 5′ end of each of these genes (e.g., GAL1-FIG2, GAL1-FLO10) at their normal chromosomal location so that the gene was under the control of the GAL1 promoter rather than its natural promoter. In Gal medium overexpression of each of these genes enhances flocculation. The order of enhancement is FLO1 > FLO10 > FLO11 > FIG2 (Fig. 1A). GAL1-FLO1 is so flocculent that the cells grow in a huge clump and remain at the bottom of the tube. GAL1-FIG2 is only slightly flocculent. This sedimentation is caused by the formation of cell aggregates. However, these cell aggregates have different characteristics. The GAL1-FLO1 and GAL1-FLO10 aggregates are calcium dependant and both are reversibly inhibited by the presence of mannose. But, unlike the GAL1-FLO1 aggregates, the GAL1-FLO10 aggregates are also inhibited by the presence of maltose, sucrose, and glucose. The GAL1-FLO11 aggregates are calcium independent and are not inhibited by the presence of these sugars. The GAL1-FIG2 flocculation is too weak to be analyzed by these methods.
Figure 1.
Overexpression of cell wall proteins causes flocculation and adherence to the agar. (A) Ten-milliliter cultures of yeast strains were grown in YPGal overnight, swirled briefly in a Vortex mixer, and photographed immediately and after 30, 60, and 90 min (numbers to the right). Yeast strains: 1, wild type (10560–2B); 2, flo11 (WY168); 3, flo11 GAL1-FIG2 (WY297); 4, GAL1-FLO11 (WY334); 5, flo11 GAL1-FLO10 (WY341); and 6, flo11 GAL1-FLO1 (WY340). (B) The same strains used in A were patched to YPD or YPGal plates. After incubation for 5 days, the plates were washed under a stream of water and rephotographed. The wild-type strain is less invasive on a YPGal plate than on a YPD plate. (C) The same strains used in A were grown in YPD or YPGal liquid medium overnight and swirled briefly in a Vortex mixer, and a fraction of the cells were spotted on an agar (2%) plate. The plate was let dry for 30 min, washed, and rephotographed. The flo11 GAL1-FLO1 strain looks denser on the unwashed plate because the cells are flocculent and stick together in large clumps.
The enhanced expression of some of these cell wall proteins also results in invasive growth. Mutations in FLO11 abolish haploid invasive growth, showing that in wild type none of the other flocculins can compensate for the flo11 defect (11, 12). To test whether other family members could function in invasive growth, we overexpressed FIG2, FLO10, or FLO1 in a flo11 mutant and assayed each strain to determine whether such heterologous overexpression could suppress the flo11 defect in haploid invasion. Both FIG2 and FLO10 overexpression cause invasion in the flo11 background, whereas FLO1 overexpression does not (Fig. 1B).
Haploid invasion may have several components, including adherence to agar and penetration of the agar. To distinguish between these we tested for the ability of various strains to adhere to agar in the absence of growth. The GAL1-promoted versions of FLO11, FLO1, FLO10, and FIG2 were first grown on Gal medium, spotted on agar that contained no nutrients, and allowed to dry. The plate was then washed under the tap as usual. Under these conditions the flo11 strain does not adhere, whereas wild type and the GAL1-promoted FLO11, FLO10, and FIG2 strains do adhere (Fig. 1C). These data suggest that penetration of the agar is not required for “haploid invasion.” Furthermore, the fact that the GAL1-FLO1 strain does not adhere to the agar but is the most flocculent strain tested suggests that adherence to agar is different from the cell–cell adherence required for flocculation. Thus, these proteins have distinct but overlapping functions: Fig2p, Flo1p, Flo10p, and Flo11p promote flocculation; Fig2p, Flo10p, and Flo11p promote both flocculation and invasion.
Adhesive Cell Wall Proteins Are Interchangeable for Mating.
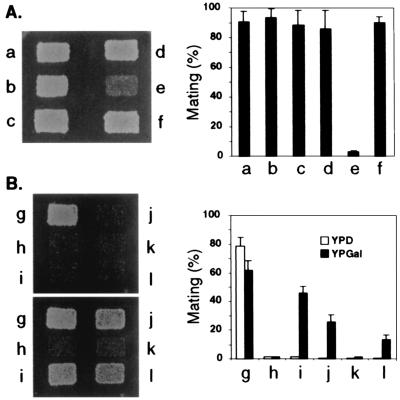
Fig2p is not expressed in a vegetative haploid cell, but it is strongly induced by exposure to pheromone (15) or to cells of the opposite mating type (data not shown). We find that Fig2p is responsible for the pheromone-induced invasive growth described by others (34) (data not shown). We also find that Fig2p is required for mating in strains that fail to produce the yeast mating agglutinins, suggesting that these proteins have a overlapping function (Fig. 2A). A MATa fig2 aga1 strain shows more than 30-fold reduction in mating as compared with a wild-type, an aga1, or a fig2 strain. We observed that Fig2p is also involved in mating in MATα strains. MATα strains express Fig2p, Aga1p, and Sag1p (16, 18). The MATα fig2 aga1, fig2 sag1, and aga1 sag1 strains mate as well as wild type, whereas a fig2 aga1 sag1 strain has a strong mating defect. These data reveal an unexpected function for these cell wall proteins: both Aga1p and Sag1p are involved in mating in MATα cells. On solid media, the Fig2p protein can bypass the requirement for the Aga1p and Sag1p proteins and vice versa.
Figure 2.
Adhesive cell wall proteins are interchangeable for mating. The images on the left are plate matings and the histograms on the right are the results of quantitative matings. (A) MATa stains were patched onto a YPD plate, grown overnight, replicated to a YPD plate, and mated with a wild-type MATα tester lawn (10560–45B). Matings were carried out for 4 h, after which the mating mixture was replicated to a selective medium (YNB) plate. Left shows growth of the diploids. Yeast strains: a, wild type (10560–31D); b, fig2 (WY362); c, aga1 (WY313); d, aga2 (WY486); e, fig2 aga1 (WY364); and f, fig2 aga2 (WY488). The same strains were also used in a quantitative mating assay (Materials and Methods) and the results are shown on the Right. The mating percentage is defined as diploids/(diploids + nonmated haploids). Bars and error bars show means of three assays and standard errors, respectively. (B) Cell surface proteins can compensate for the mating defect of a fig2 aga1 strain. A MATa fig2 aga1 flo11 strain mated 2–3 times less well than a fig2 aga1 strain, implying the involvement of Flo11p in mating. Thus the following mating tests were carried out in the fig2 aga1 flo11 background. MATa strains were mated with a wild-type MATα tester strain (10560–4B) in a plate mating assay and quantitative mating assay similar to those in A. Upper Left shows mating on a YPD plate and Lower Left on a YPGal plate. Yeast strains: g, wild type (10560–14A); h, fig2 aga1 flo11 (WY350); i, fig2 aga1 GAL1-FLO11 (WY345); j, aga1 flo11 GAL1-FIG2 (WY310); k, fig2 aga1 flo11 GAL1-FLO1 (WY367); and l, fig2 aga1 flo11 GAL1-FLO10 (WY368).
The mating defect in a MATa fig2 aga1 strain appears to be in adhesion of the cells of opposite mating type to each other. The cells show normal sensitivity to pheromone and form projections that look normal in number and appearance. However, in a mating mixture with wild type MAT α cells, there are virtually no zygotes or pairs of cells attached by their projections suggesting that the mating defect results from a failure to adhere to cells of the opposite mating type (data not shown).
Several of the vegetative flocculins can compensate for the mating defect of a fig2 aga1 mutant (Fig. 2B). The FLO10 and FLO11 genes when overexpressed can compensate for the mating defect of the fig2 aga1 strain. A GAL1-FLO11 fig2 aga1 strain induced with Gal dramatically increases the mating efficiency, indicating that Flo11p can bypass the defect created by the absence of Fig2p and Aga1p. None of those flocculins can suppress the mating defect of fig2 aga1 when expressed from their natural promoter. Thus, Fig2p, Aga1p, and Sag1p have specialized to function as mating-specific adhesins, and Flo10p and Flo11p, which mediate adhesion in other contexts, can replace Fig2p, Aga1p, and Sag1p when overexpressed.
Ectopic Expression of the Flocculins Promotes Filamentous Growth.
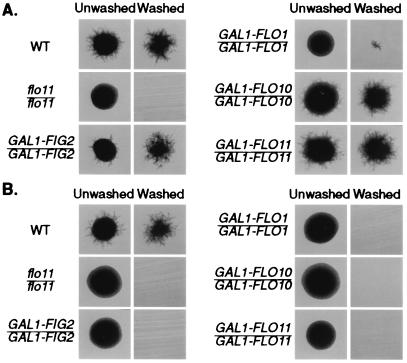
The ability of the various flocculins to promote both mating and invasion when expressed ectopically prompted a test of their ability to replace FLO11 function for filamentous growth. Filamentous growth is a property of diploids: FLO11/FLO11 strains make prolific filaments, whereas flo11/flo11 strains make smooth round colonies lacking filaments (12, 35). We expressed several of the flocculins from the GAL1 promoter and tested these constructs in flo11/flo11 diploids for their ability to promote filamentation. Remarkably, GAL1-FIG2 and GAL1-FLO10 induced prolific filaments in the absence of FLO11 (Fig. 3). Expression of GAL1-FLO1 fails to induce filaments that protrude from the colony, although there was evidence of a few filaments remaining embedded in the agar after washing. The filaments produced by GAL1-FIG2 and GAL1-FLO10 appear normal.
Figure 3.
Ectopic expression of the flocculins promotes filamentous growth. Strains expressing different flocculin genes from the GAL1 promoter were streaked to SLAR + Gal plates (A) or SLAD plates (B), grown for 4 days, and photographed before and after washing. Yeast strains: WT (WY454), flo11/flo11 (WY449), flo11/flo11 GAL1-FIG2/GAL1-FIG2 (WY452), flo11/flo11 GAL1-FLO1/GAL1-FLO1 (WY450), flo11/flo11 GAL1-FLO10/GAL1-FLO10 (WY451), and flo11/flo11 GAL1-FLO11/GAL1-FLO11 (WY415).
FLO10 Can Bypass Loss of FLO11 Function.
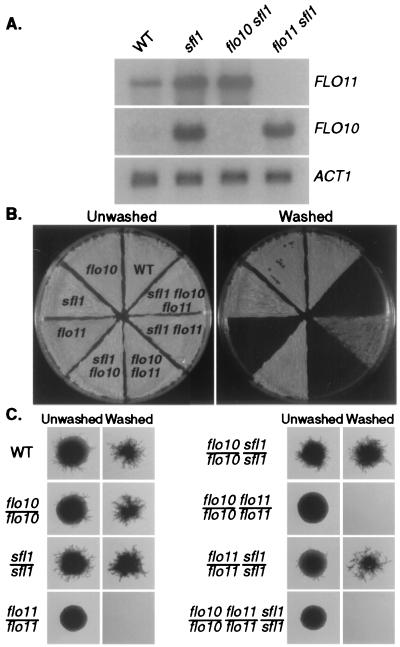
The ability of overexpression of FLO10 to bypass a defect in FLO11 raises the question of whether FLO10 under its own promoter can supply FLO11 function. Previous work had shown that deletion of the regulatory gene SFL1 partially bypasses the flo11 defect for both invasion and filamentation (36). SFL1 is known to repress the expression of some genes (36–38). Therefore, one hypothesis to explain the adhesion and filamentation phenotype of a flo11 sfl1 mutant is that FLO10 is derepressed in an sfl1 mutant and can supply the compensatory function missing in flo11. This hypothesis predicts that FLO10 is repressed by SFL1 and that FLO10 function is required for filamentation and adhesion in a sfl1 flo11 mutant.
Northern analysis shows that FLO10 expression is under SFL1 control (Fig. 4A). Both FLO11 and FLO10 messages are repressed by SFL1, but FLO10 is much more derepressed in the sfl1 mutant. To test whether derepression of FLO10 is responsible for the suppression of the flo11 defect in sfl1 strains, we compared sfl1 flo11 strains with sfl1 flo11 flo10 strains. As predicted by this repression model, sfl1 flo11 strains adhere but sfl1 flo11 flo10 strains do not (Fig. 4B). Moreover, sfl1/sfl1 flo11/flo11 diploids filament but sfl1/sfl1 flo11/flo11 flo10/flo10 diploids do not (Fig. 4C). These results are a direct demonstration that the FLO genes have interchangeable functions, even under the control of their own promoters.
Figure 4.
FLO10 can bypass loss of FLO11 function. (A) FLO10 and FLO11 expression are repressed by SFL1. Northern blots of total RNA (10 μg) prepared from strains WT (10560–2B), sfl1 (WY458), sfl1 flo10 (WY462), and sfl1 flo11 (WY466) were hybridized with FLO10, FLO11, and ACT1 probes, respectively. (B) Derepression of FLO10 leads to haploid invasion. Strains WT (10560–2B), flo10 (WY456), sfl1 (WY458), flo11 (WY460), flo10 sfl1 (WY462), flo10 flo11 (WY464), flo11 sfl1 (WY466), and flo10 flo11 sfl1 (WY468) were patched to a YPD plate. After 5 days of incubation, the plate was washed under a stream of water and rephotographed. (C) Derepression of FLO10 promotes filamentous growth. Strains WT (WY454), flo10/flo10 (WY479), sfl1/sfl1 (WY480), flo11/flo11 (WY481), flo10/flo10 sfl1/sfl1 (WY482), flo10/flo10 flo11/flo11 (WY483), flo11/flo11 sfl1/sfl1 (WY484), and flo10/flo10 flo11/flo11 sfl1/sfl1 (WY485) were streaked on a SLAD plate, grown for 7 days, and photographed before and after washing.
Adhesion Is Associated with the Presence of Flo11p and Fig2p on the Cell Surface.
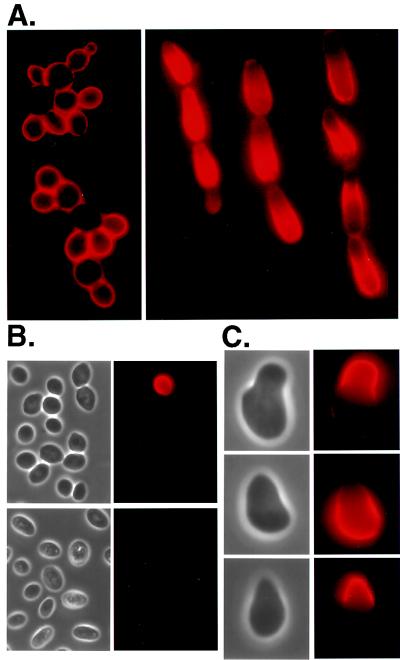
One explanation for the role of Flo11p function in haploid invasion and diploid filamentation is that this protein is localized to the cell wall, where it promotes adhesion to various surfaces. To test this idea we replaced the chromosomal copy of FLO11 with a HA-tagged version, FLO11–3HA (see Materials and Methods), and visualized the cellular location of Flo11p by indirect immunofluorescence. This tagged protein, the only copy of Flo11p in the strain, is fully functional in both haploid invasion and diploid filamentation (data not shown). Haploid cells show uniform staining of Flo11–3HA around the surface (Fig. 5A Left). Diploid yeast form cells grown on rich medium (synthetic complete) (Fig. 5B Upper) or nitrogen starvation medium (SLAD) (Fig. 5B Lower) show little or no staining on the surface. However, filamentous cells from SLAD stain intensely (Fig. 5A Right). Many cells in the filaments show asymmetric distribution of Flo11–3HA, with the most intense staining on the surface of a daughter further away from her mother. This asymmetric distribution can be seen in older cells of the filaments as well as on the tip of the growing bud at the end of the filament. However, some cells in the filament stain all over their surface. The failure of diploid yeast-form cells to stain is significant because these cells originate from the same colony as the filaments that stain intensely.
Figure 5.
Adhesion is associated with the presence of Flo11p and Fig2p on the cell surface. The Flo11p and Fig2p proteins were tagged with HA (see Materials and Methods). Yeast cells were harvested, fixed, treated with mouse anti-HA antibody, and stained with Cy3-conjugated goat anti-mouse IgG antibody. (A) Haploid yeast cells (strain WY423) grown on synthetic complete medium show uniform staining of Flo11–3HA (Left). Diploid pseudohyphal cells (strain WY453) grown on SLAD (with 0.2 mM l-histidine hydrochloride added) often show polarized staining (Right); however, some show uniform staining (see filament on left). (B) Most diploid yeast cells (WY453) fail to stain Flo11p whether they are grown on synthetic complete medium (Upper) or SLAD (Lower). (C) Haploid yeast cells treated with α factor form mating projections (Left). Only the projections show Fig2–3HA staining (strain WY427) (Top Right). Flo11–3HA appears on the body of the cell and not in the projection (strain WY423) (Middle Right). Flo11–3HA induced with Gal appears only in the projection (strain WY491) (Bottom Right).
Fig2p, in contrast, appears to be localized to the projection that forms during mating (Fig. 5C Top). A functional HA-tagged version of Fig2p (Fig2–3HA) is induced by mating pheromone and localizes to the projection, with little staining on the body of the cell. The localization of Flo11p in mating cells shows a completely different distribution from Fig2p. When haploid cells are induced with pheromone, Flo11p appears on the body of the cell but is absent from the projection (Fig. 5C Middle). The absence of Flo11p from the projection could explain why Flo11p fails to restore mating in cells lacking Fig2p and Aga1p. Presumably, it is not in the proper location to carry out this function. Since overproduction of Flo11p dramatically increased the mating efficiency of a fig2 aga1 strain (Fig. 4C), we inserted a GAL1 promoter at the 5′ end of the FLO11–3HA construct (GAL1-FLO11–3HA) to determine the localization of Flo11p when it is overproduced during mating. The GAL1-FLO11–3HA strain was treated with pheromone upon the shift from glucose to galactose media. When FLO11 is induced by galactose during pheromone treatment, the majority of the Flo11p (Flo11–3HA) is localized to the projection (Fig. 5C Bottom).
Discussion
Our data show that one FLO gene can compensate for another in diverse morphogenetic events: flocculation, mating, haploid invasion, and filamentation. This apparent functional redundancy raises the question of why mutations in any one of the genes results in a mutant phenotype. The answer is that the phenotype of even a wild-type strain depends on both the regulation and localization of these proteins. For example, FLO11 is required for both haploid invasion and diploid filamentation (11, 12) on our media. Because none of the other flocculins is expressed at sufficient levels under these conditions, the flo11 colonies have a distinct mutant phenotype. However, when overexpressed, FIG2 or FLO10 can bypass the requirement for FLO11 for both filamentation and haploid invasion.
In wild-type cells, the FIG2 gene is expressed only when it is induced by mating pheromone (15), and FLO10 is not produced in sufficient quantities under these conditions to bypass the flo11 defect. However, when repression of FLO10 is relieved (in the sfl1 mutant), it is fully capable of supplying a compensatory function for haploid invasion and diploid filamentation. Therefore, the distinction among FIG2, FLO10, and FLO11 in vegetative cells occurs at the level of the cis-acting promoters specific to each gene and the trans-acting regulators that control them.
Localization Is Also Responsible for the Diversity of Function.
In haploid cells grown on rich medium Flo11p is expressed at high levels and uniformly distributed on the cell surface. These features explain the requirement for Flo11p for adhesion of haploid cells to the agar surface. Diploids grown on rich medium express very little Flo11p on their cell surface, which is consistent with their reduced agar adhesion compared with haploids. On nitrogen starvation media most of the diploid cells in a colony are in the yeast form and not in filaments. Yet, only the filamentous cells express Flo11p. The presence of Flo11p at the cell surface in filamentous cells but not yeast-form cells within the same colony is consistent with the requirement of Flo11p for filamentous growth. Although flo11/flo11 diploids fail to form filaments, they do make long cells in response to nitrogen starvation, suggesting that the formation of long cells per se is not sufficient to form filaments.
The localized deposition of Flo11p at the site of bud formation could prevent separation of mother cells from their daughters. Indeed, our data show polarization of Flo11p to the growing cell tip in the filaments. However, not all cells in the filament show this polarization; some cells have Flo11p distributed over their entire surface. As with other secreted proteins, Flo11p may be at first concentrated toward the bud tip, but subsequently equilibrated over the entire surface. Flo11p could also be required on the cell surface to adhere the mothers and newly born daughters to the agar surface and to each other. According to this model, the other events induced by nutritional starvation—alteration in cell cycle, elongation of the cells, and unipolar budding—would be responsible for the directional growth of an emerging filament.
The induction of Fig2p by mating pheromone and its localization to the mating projection is consistent with its role in cell–cell adherence, which is required as a prelude to cell fusion in mating. In support of this model, Fig2p is required in an aga1 flo11 strain to permit adherence and fusion. This raises the question of why Flo11p cannot provide this adhesion function in the fig2 aga1 strain. Our data suggest that Flo11p cannot normally function in mating because it is not present in the projection. Fig2p is induced to very high levels by mating pheromone, whereas Flo11p shows no induction by pheromone (35). Presumably, the newly synthesized Fig2p is secreted in a polarized manner to the growing projection, where it is poised to promote adhesion with cells of the opposite mating type. Only when Flo11p is overexpressed by Gal during pheromone induction can it localize to the projection and function in mating. Thus, both regulation and localization control the functional distinction between these two proteins.
Evolution of Diversity Among the Flocculins.
The diversity in adhesive properties among the flocculins with similar domain structures suggests that recombination could generate new flocculins with novel adhesive properties. Previous studies have demonstrated the generation of a novel flocculin by recombination between FLO5 on chromosome VIII and a flocculin pseudogene YAL065 on chromosome I (39). This mechanism of creating diversity in surface antigens by recombination would be similar to that used by other pathogenic microorganisms (40, 41).
Supplementary Material
Acknowledgments
We thank Scott Erdman and Michael Snyder for providing yeast strains. We thank David Sabatini, Mike Lorenz, Tim Galitski, and Adrian Halme for their critical readings of the manuscript and members of the Fink laboratory for helpful discussions. We also thank Nicki Watson for assistance in microscopy. B.G. is supported by a National Research Service Award (F32GM19181–02). Q.F. is supported by a Damon Runyon Postdoctoral Fellowship (DRG-1418). G.R.F. is an American Cancer Society Professor of Genetics.
Abbreviation
- HA
hemagglutinin epitope
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220420397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220420397
References
- 1.Hostetter M K. Clin Microbiol Rev. 1994;7:29–42. doi: 10.1128/cmr.7.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundstrom P. Curr Opin Microbiol. 1999;2:353–357. doi: 10.1016/S1369-5274(99)80062-9. [DOI] [PubMed] [Google Scholar]
- 3.Hoyer L L, Scherer S, Shatzman A R, Livi G P. Mol Microbiol. 1995;15:39–54. doi: 10.1111/j.1365-2958.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]
- 4.Fu Y, Rieg G, Fonzi W A, Belanger P H, Edwards J E, Jr, Filler S G. Infect Immun. 1998;66:1783–1786. doi: 10.1128/iai.66.4.1783-1786.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cormack B P, Ghori N, Falkow S. Science. 1999;285:578–582. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- 6.Caro L H, Tettelin H, Vossen J H, Ram A F, van den Ende H, Klis F M. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Teunissen A W, Steensma H Y. Yeast. 1995;11:1001–1013. doi: 10.1002/yea.320111102. [DOI] [PubMed] [Google Scholar]
- 8.Johnston J R, Reader H P. In: Yeast Genetics, Fundamental and Applied Aspects. Spencer J F T, Spencer D M, Smith A R W, editors. New York: Springer; 1983. pp. 205–224. [Google Scholar]
- 9.Teunissen A W, van den Berg J A, Steensma H Y. Yeast. 1995;11:735–745. doi: 10.1002/yea.320110805. [DOI] [PubMed] [Google Scholar]
- 10.Teunissen A W R H. Ph.D. Thesis. Leiden, The Netherlands: Leiden University; 1995. [Google Scholar]
- 11.Lambrechts M G, Bauer F F, Marmur J, Pretorius I S. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo W S, Dranginis A M. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 14.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 15.Erdman S, Lin L, Malczynski M, Snyder M. J Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy A, Lu C F, Marykwas D L, Lipke P N, Kurjan J. Mol Cell Biol. 1991;11:4196–4206. doi: 10.1128/mcb.11.8.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cappellaro C, Hauser K, Mrsa V, Watzele M, Watzele G, Gruber C, Tanner W. EMBO J. 1991;10:4081–4088. doi: 10.1002/j.1460-2075.1991.tb04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipke P N, Wojciechowicz D, Kurjan J. Mol Cell Biol. 1989;9:3155–3165. doi: 10.1128/mcb.9.8.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bony M, Barre P, Blondin B. Yeast. 1998;14:25–35. doi: 10.1002/(SICI)1097-0061(19980115)14:1<25::AID-YEA197>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 20.Callija G B. In: The Yeasts. Rose A H, Harrison J S, editors. Vol. 2. London: Academic; 1987. pp. 165–238. [Google Scholar]
- 21.Rose A H. In: Microbial Adhesion and Aggregation. Marshall K C, editor. Berlin: Springer; 1984. pp. 323–335. [Google Scholar]
- 22.Cross F, Hartwell L H, Jackson C, Konopka J B. Annu Rev Cell Biol. 1988;4:429–457. doi: 10.1146/annurev.cb.04.110188.002241. [DOI] [PubMed] [Google Scholar]
- 23.Sprague G F J, Thorner J. In: The Molecular Biology of the Yeast Saccharomyces. Broach J R, Pringle J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 657–744. [Google Scholar]
- 24.Kurjan J. Annu Rev Genet. 1993;27:147–179. doi: 10.1146/annurev.ge.27.120193.001051. [DOI] [PubMed] [Google Scholar]
- 25.Herskowitz I. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 26.Grenson M, Mousset M, Wiame J M, Bechet J. Biochim Biophys Acta. 1966;127:325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Styles C A, Fink G R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 28.Sherman F, Fink G R, Hicks J. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 29.Longtine M S, McKenzie A, 3rd, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 31.Boeke J D, La Croute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 32.Sprague G F., Jr Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- 33.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. NY: Green/Wiley-Interscience; 1987. [Google Scholar]
- 34.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, et al. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 35.Rupp S, Summers E, Lo H J, Madhani H, Fink G. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson L S, Fink G R. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song W, Carlson M. EMBO J. 1998;17:5757–5765. doi: 10.1093/emboj/17.19.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonouchi A, Fujita A, Kuhara S. J Biochem (Tokyo) 1994;115:683–688. doi: 10.1093/oxfordjournals.jbchem.a124396. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi O, Hayashi N, Kuroki R, Sone H. J Bacteriol. 1998;180:6503–6510. doi: 10.1128/jb.180.24.6503-6510.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudenko G, Cross M, Borst P. Trends Microbiol. 1998;6:113–116. doi: 10.1016/s0966-842x(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 41.Seifert H S. Mol Microbiol. 1996;21:433–440. doi: 10.1111/j.1365-2958.1996.tb02552.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.