Abstract
We have prepared a monoclonal Buckminsterfullerene specific antibody and report the sequences of its light and heavy chains. We also show, by x-ray crystallographic analysis of the Fab fragment and by model building, that the fullerene binding site is formed by the interface of the antibody light and heavy chains. Shape-complementary clustering of hydrophobic amino acids, several of which participate in putative stacking interactions with fullerene, form the binding site. Moreover, an induced fit mechanism appears to participate in the fullerene binding process. Affinity of the antibody–fullerene complex is 22 nM as measured by competitive binding. These findings should be applicable not only to the use of antibodies to assay and direct potential fullerene-based drug design but could also lead to new methodologies for the production of fullerene derivatives and nanotubes as well.
Until 1985, only two allotropic forms of elemental carbon were known: graphite and diamond. In 1985, a third allotrope was discovered and dubbed Buckminsterfullerene because of its geodesic character (1). The preparation of fullerenes in workable quantities led first to applications in chemical and engineering processes and subsequently to the suggested use of fullerenes in biological applications (2–4) and as templates for the design of experimental pharmaceutical agents, including those with anti-viral (5–9), antioxidant (8–12), chemotactic (13), and neuroprotective (14) activities.
We have previously demonstrated that the mouse immune repertoire is diverse enough to recognize and produce antibodies specific for fullerenes (15). We have now succeeded in isolating several monoclonal anti-C60 antibodies. The monoclonal antibody under study (1-10F-A8) is an IgG1 kappa and was prepared by standard procedures. Specificity for the C60 fullerene was determined by competitive inhibition. The sequences of the light and heavy chains were determined, and the three-dimensional structure of the Fab fragment was solved and refined by x-ray crystallographic techniques to a resolution of 2.25 Å. Finally, we have identified and modeled the probable binding site for C60 fullerene and discuss the interatomic interactions that stabilize the antibody–fullerene complex.
Materials and Methods
Production, Purification, and Crystallization of Fab.
BALB/c mice were immunized and boosted as described previously (15). Five days before the fusion, the mice were injected i.p. with 0.5 mg of the immunogen in PBS. Spleen cells were harvested and fused with the nonsecreting myeloma P3x63Ag.8.653 according to the procedure of Sharon et al. (16) Screening was by ELISA using the rabbit serum albumin (RSA) conjugate of the fullerene used in the immunization procedure. The antibody selected was designated 1-10F-8A and is an IgG1 kappa. Large-scale production of the antibody was carried out in a Cellmax System (Cellco, Kensington, MD) using a protein-free medium (Sigma S-2897).
IgG was precipitated by ammonium sulfate, dissolved in and dialyzed against PBS (150 mM NaCl/10 mM sodium phosphate, pH 7.4). Fab was prepared by papain digestion. Fab and Fc fragments and undigested IgG were separated by anion-exchange using a MonoQ column in an FPLC apparatus (Amersham Pharmacia) using a linear gradient of buffer B (50 mM Tris/1.0 M NaCl, pH 8.5). The Fab fragment eluted as a sharp peak. Fab was concentrated and desalted in Microcon-30 centrifugal concentrators (Amicon). Final solution contained Fab at a concentration of 7 mg/ml in 10 mM Tris, pH 7.0.
Crystals were grown at room temperature by the hanging-drop vapor-diffusion method using Linbro multiwell plates. Drops containing 2 μl of the protein solution and 2 μl of precipitant buffer were equilibrated against 500 μl of precipitant buffer. The reservoir contained 10% PEG 8000, 0.1 M citrate, pH 5.0. Crystals shaped as long rectangular prisms appeared after 4 days and grew to maximum size (0.8 × 0.05 × 0.05 mm) in 10–14 days.
Data Collection and Model Refinement.
Rotating anode diffraction data were collected at a temperature of 100 K, in 1° oscillation steps on a MAR 345 imaging plate and processed with the HKL suite of programs (17). Data indicate that the crystals are monoclinic, space group P21 with cell parameters a = 57.9, b = 65.75, c = 65.56, β = 112.45. Statistics for the data are listed in Table 1. The structure of the anti-fullerene Fab was solved by molecular replacement procedures using the program suite CNSSOLVE (18). For the test model, an anti-lysozyme mouse Fab (D44.1, IgG1 κ, Protein Data Bank code 1MLB) was chosen because of identical hypervariable segment lengths in the light chain complementarity-determining regions (CDRs) and H1, H2 of the heavy chain, to the anti-fullerene antibody. Best rotation and translation solutions were found with the model modified by a −20° change in the elbow angle. Single rotation and translation solutions were found (15-4 Å, F > 2σ data) with a resulting Patterson correlation coefficient of 0.3 (next highest, 0.15). A cycle of rigid body refinement, followed by a minimization refinement (6-2.25 Å, F > 2σ data) produced a model with an R-factor of 0.24 (Rfree = .29). Refinement was continued by alternate cycles of manual model building using the visual program TURBO (19) and simulated annealing refinement using CNSSOLVE. Inspection of 2Fo − Fc and Fo − Fc electron density maps clearly indicated the differences in amino acid sequence between a model and anti-fullerene Fab, and by the 5th cycle of refinement, the complete sequence was fit to electron density. Refinement continued for an additional four cycles with improvements to stereochemistry of the polypeptide and the addition of solvent water molecules. Refinement was terminated when no peaks in an Fo − Fc difference Fourier were greater than 2.5σ. Statistics for the refinement are summarized in Table 1.
Table 1.
Data collection and refinement
| Data collection | |
| Resolution limit (Å) | 2.25 |
| Data completion (I/σ[I] > 0) | 90.0% |
| Rsym (all reflections, 2 crystals) | 9% |
| Refinement | |
| Resolution (Å) | 60.00-2.25 |
| Number of reflections (F/óF > 1) | 17,688 |
| Number of protein atoms | 3,234 |
| Number of water molecules | 282 |
| Final R-factor/R-free | 18./24. |
| B-factors (Å2) main/side chain | 22.2/23.1 |
| rms deviations from ideal | |
| Bonds (Å) | 0.018 |
| Angles (degrees) | 2.106 |
Assay of Antibody by ELISA.
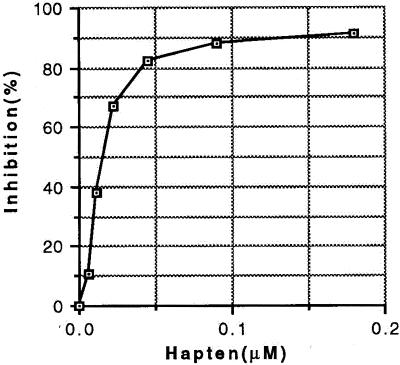
A 96-well plate (Falcon 3915) was coated with 100 μl of a solution containing 0.25 μg/ml antibody in 0.1 M sodium bicarbonate, pH 9.3, at 4°C overnight and then washed with PBS/0.1% Tween 20 three times. To solutions of biotinylated C60-rabbit serum albumin, variable amounts of a water-soluble fullerene derivative in PBS/0.1% Tween 20 were added. One-hundred microliters of each mixture was transferred to the coated 96-well plate, followed by incubation at 37°C for 2 h. After three washes with PBS/0.1% Tween 20, 100 μl of a 1:1,000 dilution of Extravidin-peroxidase (Sigma E-2886) was added and allowed to incubate at 37°C for 30 min. After washing four times with PBS/0.1% Tween 20, development was with 100 μl of o-phenylenediamine (7 mg/ml in 0.1 M citrate-phosphate buffer, pH 5.0, containing 5 μl of 30% hydrogen peroxide). Readings were made at 490 nm (Fig. 1).
Figure 1.
Specificity of monoclonal anti-fullerene antibody 11-10F-A8. Inhibitor is a water-soluble C60 fullerene derivative designated C151H134N8O48; molecular weight, 2828.74. Apparent binding constant of antibody is 22 nM based on concentration at 50% binding.
Modeling of the C60–Antibody Binding Interaction.
A model of C60 fullerene was placed at the opening of the channel to the putative binding site, and the system was minimized holding the antibody structure fixed to rms = 0.001 Å (INSIGHT II, MSI Corp., version 98.0; force field = cvff; energy, −28.2344 kcal/mol). Next, an unfixed model (rms = 0.01 Å) was constructed by allowing all atoms to move followed by molecular dynamics calculations (298 K for 5,000 steps; rms = 0.03 Å; energy, −53.9317 kcal/mol). At this point, we also made manual rotations of amino acid side chains to open the channel to the spherical pocket identified by accessible surface area calculation. After these rotations were completed, the C60 molecule was manually moved through the opening to the binding site cavity. Minimization was continued with all atoms allowed to move, i.e., unfixed, leading to a model with the C60 molecule and anti-C60 Fab fragment minimized to rms = 0.001 (energy, −120.799 kcal/mol).
Sequencing and Germ-Line Gene Assignments.
Sequencing of the variable regions of 1–10F-8A was carried out as described (20) with the following changes: total RNA was prepared using the TRIzol reagent (GIBCO/BRL). The first strand cDNA preparations used the random hexamers of the universal riboclone cDNA synthesis system (Promega). PCR was carried out as previously described (20).
Germ-line gene segments were assigned by nucleotide sequence comparisons with those of the closest germ-line genes found using the BLAST algorithm (21). For the case of the variable region of the heavy chain, where no germ-line gene was detected by nucleotide sequence analysis, putative germ-line mutations were assigned by comparison of the amino acid differences in the 24 sequences sharing 85% or greater homology with the anti-fullerene sequence.
Results and Discussion
Crystal Structure Analysis.
The polypeptides of the heavy and light chains are well defined by electron density except for residues 131 through 135 in the Fab heavy chain constant domain (CH1) that show no electron density due to apparent conformational disorder. Analysis of conformation by a Ramachandran plot shows close clustering of the main-chain torsion angles in the energetically favorable regions and identifies only one non-glycyl residue in a region generally considered as disallowed. This residue, light chain Thr-51 at the apex of CDR L2, a γ turn, is frequently noted as occurring in a high-energy region in many Ig structures. Analysis of the main-chain conformation of the L1, L2, L3, H1, and H2 hypervariable regions by sequence and three-dimensional structure comparison with other immunoglobulins confirms that standard conformations based on hypervariable loop length and sequence (i.e., canonical structure) are maintained. Thus, molecular recognition of the fullerene–RSA conjugate is accomplished by the structural motifs used by all immunoglobulins to recognize antigen or hapten–protein conjugates. The anti-fullerene antibody, however, has been constructed with a very short CDR H3 comprising only four residues (Ser, Ser, Ala, Tyr) linking framework regions. A search of all Ig structures deposited in the Protein Data Bank revealed no other antibody with a CDR H3 of four residues and only one (ID code 1IIF, an anti-peptide) containing a three-residue CDR H3.
Identification and Modeling of C60 Binding.
Identification of the C60 binding site in the anti-fullerene antibody was accomplished by accessible surface area calculation and docking/energy minimization procedures. Accessible surface area calculation with a 1.7 Å radius probe identifies a spherical-shaped cavity in the VL–VH interface delineated by the side chains of VL CDR residues Tyr-36, Gln-89, Phe-96, and by VL framework residue Phe-98, by the side chains of VH CDR residue Asn-35 and VH framework residues Val-37, Trp-47, and Trp-106 (Fig. 2). In addition, the main chain atoms of VH residues Ala-97, Thr-98, and Ser-99 (in CDR H3) also define the cavity. The cavity contains two solvent water molecules that form a solvent bridge between the side chains of VL Gln-89 and VH Asn-35.
Figure 2.
Proposed C60 binding site identified by solvent-accessible surface calculation. Dot representation of the accessible surface was derived from the program MS (Michael Connolly, University of California, San Francisco) using a 1.7-Å radius probe. The indicated cavity is 7 Å maximum in width and lies in the VL–VH interface.
Accessibility to the fullerene binding site is via a 4.7-Å wide channel fashioned by the CDR L3 and the short CDR H3 hypervariable loop. The cavity identified in the VL–VH interface is approximately 7 Å in diameter, too small for a C60 fullerene of 10-Å diameter, incorporating the van der Waals radii of the carbon atoms. Nonetheless, manual docking of a model C60 into the cavity and using an energy minimization protocol results in relaxation of the surrounding Fab side chains and 11o rotation of the VL and VH domains (Fig. 3). The antibody surface area buried by the VL–VH interface is a relatively small 1,100 Å2, consistent with other anti-hapten antibodies that undergo large relative VL–VH displacements upon hapten binding (22).
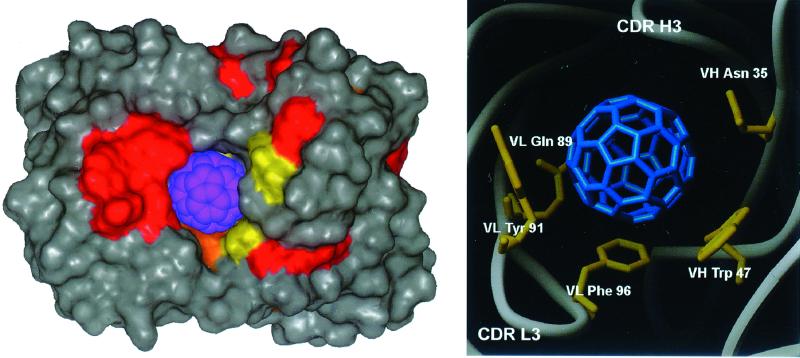
Figure 3.
Model of C60 binding site. (Left) Van der Waals surface representation of C60 bound to the anti-fullerene antibody. VL domain is to the left, VH to the right. Surface is color-coded by amino acid: red, tyrosine; orange, phenylalanine; yellow, tryptophan. All other amino acids are colored gray; C60 is colored blue. (Right) Residues involved in stacking interactions to modeled C60. View orientation is same as Left: VL, left; VH, right.
In addition to confirming that the fullerene binding site is the spherical cavity identified by accessible surface area calculation, the modeling and energy minimization of the Fab–C60 complex suggests several pi–bond stacking interactions between the fullerene and the antibody. Aromatic side chains of VH Trp-47, VL Tyr-91, and VL Phe-96 as well as side chains for VH Asn-35 and VL Gln-89 lie parallel to the C60 molecule (Fig. 3). In addition, the potential for a weak hydrogen bond from the VL Tyr-36 hydroxyl to the fullerene (3.15 Å) is also noted. Stacking interactions to C60 have been previously described in the crystal structure of C60 solvated by benzene (23). The solvent benzene was noted to lie over the electron-rich C60 pentagon–pentagon bonds, clearly establishing the nature of the stacking interaction. Pi-system stacking interactions are well established in x-ray crystal structures of antibody–antigen complexes. For example, in the antibody–antigen complex of the anti-lysozyme antibody D44 (24), the stacking interaction involved an aromatic side chain, tryptophan, from the antibody and an arginine from the antigen. It is not surprising, therefore, that the anti-fullerene antibody uses similar stacking interactions for the binding of fullerene.
Fullerenes have an extensive and well characterized resonance structure (25); as such, weak hydrogen bond interactions between the fullerene and antibody could marginally contribute to the stability of the antibody–fullerene complex. As an example, the anti-lysozyme antibody D1.3 uses a tyrosine phenyl ring in the antibody as a hydrogen bond acceptor for a glutamine side chain in the antigen (26). Whereas the C60 molecule is not aromatic in nature, localized double bonds may act as a weak hydrogen bond acceptor with the hydroxyl of VL Tyr-36.
It should be noted that the stacking and hydrogen bond interactions to the fullerene would be weak interactions. Hydrophobic desolvation effects will dominate the thermodynamics of the antibody–fullerene interaction. The fullerene surface area buried by the antibody is 337 Å2, 90% of the entire C60 surface area, indicative of the deep binding of the fullerene in the VL–VH interface. The surface area of interaction coupled with the measured 22 nM binding constant for the fullerene–antibody interaction translates to approximately 22 cal/M/Å2 for hydrophobic interaction, quite in line with the contribution of hydrophobic surface area to the free energy of antibody–antigen interactions (27). Complete description of the precise geometry of the fullerene–protein interaction must, however, wait until we have successfully cocrystallized the antibody–C60 complex.
Sequence Analysis.
Nucleotide sequence comparisons have identified the potential germ-line genes for the VL and VJ regions and reveal no somatic mutations for amino acids located in the fullerene binding site. For the VL domain, there seems to be no ambiguity in assignment of the germ-line with an identity of 95% for the nucleotides encoding the Vk10 subgroup. Homology with the nucleotides coding the J4 joining region is 100% with no frame reading errors in the VJ junction. For the VH domain, the nucleotide sequence search identified no potential germ-line genes. Likewise, a BLAST search of the amino acid sequence of residues 1 through 97 detected no amino acid sequence homology exceeding 88%. Nonetheless, for the 24 sequences for heavy chain amino acids in the comparison (homologies >85%), a consensus of 11 amino acid differences to the anti-fullerene antibody were noted, with none occurring at the ligand binding site (Fig. 4). This result is in agreement with hapten-recognizing features of other antibodies showing severe restriction of germ-line gene usage. In one important instance, restriction of germ-line gene usage has been proposed to ensure high avidities for antigen in the early immune response (28). The lack of somatic mutation of fullerene binding residues, however, does not rule out affinity maturation of the anti-fullerene antibody. Hawkins et al. (29) postulate that improvement in affinity must be mediated by altered residues at the periphery of the combining site, or in framework residues, by direct interactions. Although it is difficult to envision that the humoral immune system has indeed evolved specific recognition for such an insoluble molecule as fullerene, it is nonetheless clear that the immune repertoire is diverse enough to recognize the unusual structure of fullerene. The binding interactions noted by the structural and modeling studies of the anti-fullerene antibody will represent binding modes for the biomolecular recognition of fullerene-based molecules.
Figure 4.
VL and VH amino acid sequence alignment and somatic mutations of germ-line genes. CDRs (Kabat definitions) in bold; residues with side chains contacting fullerene are underlined.
The antibody molecule has been widely presented as a paradigm of biological individuality; functional and structural individuality and specificity is superimposed on a carefully conserved structure. With such an understanding of the antibody molecule, there is a continuing search for new uses of antibodies in regulating immune responses, in mimicking foreign antigens, in quantitating and localizing particular molecules, and in delivering therapeutic agents. The production and structural analysis of the anti-fullerene antibody has important consequences for the measurement of dosage and serum levels of fullerene derivatives of pharmacological use. Moreover, fullerene is highly modifiable with the addition of side chains to transform the molecule's solubility and functional characteristics. As such, it may be possible to modulate antibody/antigen recognition by covalent modification of antibody-bound fullerene molecules, thereby increasing antibody/antigen contact surface area or idealizing hydrogen bonds or ionic interactions. Thus, as antibodies are nature's customizable molecule, fullerenes may become the immunologist's customizable tool.
Acknowledgments
This research was supported by grants from the National Aeronautics and Space Administration/Model Institutes for Excellence (to B.C.B.), and the National Institutes of Health and C60, Inc., Toronto (to B.F.E.).
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1EMT).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210396197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210396197
References
- 1.Kroto H W, Heath J R, O'Brien S C, Curl R F, Smalley R E. Nature (London) 1985;318:162–163. [Google Scholar]
- 2.Jensen A W, Wilson S R, Schuster D I. Bioorg Med Chem. 1996;4:767–779. doi: 10.1016/0968-0896(96)00081-8. [DOI] [PubMed] [Google Scholar]
- 3.Da Ros T, Prato M. J Chem Soc Chem Commun. 1999;1999:663–669. [Google Scholar]
- 4.Wilson S R. In: The Fullerene Handbook. Kadish K, Ruoff R, editors. New York: Wiley; 2000. , in press. [Google Scholar]
- 5.Sijbesma R, Srdanov G, Wudl F, Castoro J A, Wilkins C, Friedman S H, DeCamp D L, Kenyon G L. J Am Chem Soc. 1993;115:6510–6512. [Google Scholar]
- 6.Schinazi R F, Sijbesma R, Srdanov G, Hill C L, Wudl F. Antimicrob Agents Chemother. 1993;37:1707–1710. doi: 10.1128/aac.37.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman S H, Ganapathi P S, Rubin Y, Kenyon G L. J Med Chem. 1998;41:2424–2429. doi: 10.1021/jm970689r. [DOI] [PubMed] [Google Scholar]
- 8.Chiang L Y, Lu J T, Lin J T. J Chem Soc Chem Commun. 1995;1995:1283–1284. [Google Scholar]
- 9.Lu L-H, Lee Y-T, Chen H-W, Chiang L Y, Huang H-C. Br J Pharmacol. 1998;123:1097–1102. doi: 10.1038/sj.bjp.0701722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straface E, Natalini B, Monti D, Franceschi C, Schettini G, Bisaglia M, Fumelli C, Pincelli C, Pellicciari R, Malorni W. FEBS Lett. 1999;454:335–340. doi: 10.1016/s0014-5793(99)00812-1. [DOI] [PubMed] [Google Scholar]
- 11.Lin A M, Chyi B Y, Wang S D, Yu H H, Kanakamma P P, Luh T Y, Chou C K, Ho L T. J Neurochem. 1999;72:1634–1640. doi: 10.1046/j.1471-4159.1999.721634.x. [DOI] [PubMed] [Google Scholar]
- 12.Chueh S C, Lai M K, Lee M S, Chiang L Y, Ho T I, Chen S C. Transplant Proc. 1999;31:1976–1977. doi: 10.1016/s0041-1345(99)00234-1. [DOI] [PubMed] [Google Scholar]
- 13.Toniolo C, Bianco A, Maggini M, Scorrano G, Prato M, Marastoni M, Tomatis R, Spisani S, Palu G, Blair E D. J Med Chem. 1994;37:4558–4562. doi: 10.1021/jm00052a015. [DOI] [PubMed] [Google Scholar]
- 14.Dugan L L, Turetsky D M, Du C, Lobner D, Wheeler M, Almli C R, Shen C K, Luh T Y, Choi D W, Lin T S. Proc Natl Acad Sci USA. 1997;94:9434–9439. doi: 10.1073/pnas.94.17.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B X, Wilson S R, Das M, Coughlin D J, Erlanger B F. Proc Natl Acad Sci USA. 1998;95:10809–10813. doi: 10.1073/pnas.95.18.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharon J, Morrison S L, Kabat E A. Proc Natl Acad Sci USA. 1979;76:1420–1424. doi: 10.1073/pnas.76.3.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Brünger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Roussel, A. & Inisan, A. G. (1992) turbo-frodo. Technopole de Chateau-Gombert, Europarc Bat. C. Marseille, France.
- 20.Leu J-G, Chen B X, Diamanduros A W, Erlanger B F. Proc Natl Acad Sci USA. 1994;91:10690–10694. doi: 10.1073/pnas.91.22.10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) 215, 403–410. [DOI] [PubMed]
- 22.Stanfield R L, Takimoto-Kamimura M, Rini J M, Profy A T, Wilson I A. Structure. 1993;1:83–93. doi: 10.1016/0969-2126(93)90024-b. [DOI] [PubMed] [Google Scholar]
- 23.Meidine M F, Hitchcock P B, Kroto H W, Taylor R, Walton D R. J Chem Soc Chem Commun. 1992;1992:1534–1537. [Google Scholar]
- 24.Braden B C, Souchon H, Eisele J-L, Bentley G A, Bhat T N, Navaza J, Poljak R J. J Mol Biol. 1994;243:767–781. doi: 10.1016/0022-2836(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 25.Klein D J, Schmalz T G, Hite G E, Seitz W A. J Am Chem Soc. 1986;108:1301–1302. [Google Scholar]
- 26.Bhat T N, Bentley G A, Fischmann T O, Boulot G, Poljak R J. Nature (London) 1990;347:483–485. doi: 10.1038/347483a0. [DOI] [PubMed] [Google Scholar]
- 27.Ysern X, Fields B A, Bhat T N, Goldbaum F A, Dall'Acqua W, Schwarz F P, Poljak R J, Mariuzza R A. J Mol Biol. 1994;238:496–500. doi: 10.1006/jmbi.1994.1309. [DOI] [PubMed] [Google Scholar]
- 28.Roost H P, Bachmann M F, Haag A, Kalinke U, Pliska V, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 1995;92:1257–1261. doi: 10.1073/pnas.92.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkins R E, Russell S J, Baier M, Winter G. J Mol Biol. 1993;234:958–964. doi: 10.1006/jmbi.1993.1650. [DOI] [PubMed] [Google Scholar]