Abstract
A prostate-specific gene, PCGEM1, was identified by differential display analysis of paired normal and prostate cancer tissues. Multiple tissue Northern blot analysis revealed that PCGEM1 was expressed exclusively in human prostate tissue. Analysis of PCGEM1 expression in matched normal and primary tumor specimens revealed tumor-associated overexpression in 84% of patients with prostate cancer by in situ hybridization assay and in 56% of patients by reverse transcription–PCR assay. Among various prostate cancer cell lines analyzed, PCGEM1 expression was detected only in the androgen receptor-positive cell line LNCaP. Extensive DNA sequence analysis of the PCGEM1 cDNA and genomic DNA revealed that PCGEM1 lacks protein-coding capacity and suggests that it may belong to an emerging class of noncoding RNAs, also called “riboregulators.” The PCGEM1 locus was mapped to chromosome 2q32. Taken together, the remarkable prostate-tissue specificity and androgen-dependent expression of PCGEM1 as well as its elevated expression in a significant percentage of tumor tissues suggest specific functions of PCGEM1 in the biology and tumorigenesis of the prostate gland.
Keywords: riboregulator, differential display, androgen regulation, noncoding RNA
Prostate cancer (CaP) is the most common malignancy in men in the United States and the second leading cause of cancer mortality (1). The serum prostate-specific antigen (PSA) test has revolutionized the early detection of CaP (2). Because of the prostatic epithelial-cell-specific expression of PSA, it is also of value as a biomarker for disease remission/progression after treatment (2). Although PSA is effective in identifying men who may have CaP, it is often elevated in men with benign prostatic hyperplasia, prostatitis, and other nonmalignant disorders (2). Therefore, identification of additional CaP-specific molecular markers is needed to refine the diagnosis as well as prognosis for CaP. It is also recognized that early detection of CaP presents challenges with respect to predicting the clinical course of disease for individual patients (2). The wide spectrum of biologic behavior exhibited by prostatic neoplasms calls for the identification of biomarkers that may be able to distinguish a slow growing cancer from a more aggressive cancer with a potential to metastasize (2). CaP-associated molecular genetic alterations are being unraveled by using various strategies involving: (i) analyses of genes commonly involved in human cancer, (ii) positional cloning of putative genes on frequently affected chromosome loci in CaP, and (iii) gene expression profiling of normal and tumor specimens of patients with CaP (3).
The discovery of additional prostate-specific genes has also resulted in enthusiasm for evaluating their potential in CaP diagnosis and disease progression. Increased expression of prostate-specific membrane antigen (PSMA) has been correlated with more aggressive CaP (4). CaP-associated expression of PSMA is being evaluated for imaging of CaP by radiolabeled anti-PSMA monoclonal antibodies (4). Furthermore, promising immunotherapy approaches are being pursued with PSMA peptides (5). Recently described prostate-specific genes include an androgen-regulated homeobox gene, NKX3.1, which exhibits function(s) in mouse prostate growth/development (6, 7) and shows overexpression in a subset of CaP (26). A prostate-specific gene, prostate stem cell antigen, has been shown to exhibit overexpression in CaP (8). The prostate-specific/androgen-regulated serine proteases, prostase and TMPRSS2 (9, 10), and a prostate-specific gene, DD3 (11), represent the latest additions to a small number of reports describing the discovery of prostate-specific genes.
In this report, we describe the discovery of a prostate-specific gene, PCGEM1, which was identified during our characterization of CaP-associated gene expression alterations. The most striking aspect of PCGEM1 characterization represents its prostate-tissue-specific expression with restricted expression in glandular epithelial cells. Computational analysis of PCGEM1 cDNA sequence revealed unexpected characteristics that suggest that PCGEM1 does not code for a protein, a property similar to a recently described prostate-specific gene, DD3 (11). Thus, PCGEM1 and DD3 may define a previously uncharacterized class of prostate-tissue-specific genes whose functions remain to be determined. Elevated PCGEM1 expression in a significant fraction of CaP specimens further suggests the role of PCGEM1 overexpression in prostate epithelial cell proliferation and/or tumorigenesis.
Materials and Methods
CaP Specimens and Human CaP Cell Lines.
Matched CaP and adjacent normal prostate tissues were obtained from patients who had undergone radical prostatectomy at Walter Reed Army Medical Center. Tissue histopathology and microdissections were performed as described (12). LNCaP, DU145, and PC-3 CaP cell lines were obtained from American Type Culture Collection. DuPro-1 is a nude mice xenograft (13). CPDR-1 is a primary CaP-derived cell line immortalized by retroviral vector LXSN 16 E6 E7 expressing the E6 and E7 genes of the human papilloma virus 16 (a gift from D. Galloway, Fred Hutchinson Cancer Research Center, Seattle; L.D. and S.S., unpublished work).
Preparation of RNA and Differential Display (DD) Analysis.
Total RNA was prepared from Optimal Cutting Temperature Compound (Miles, Diagnostics Division, Elkhart, IN) embedded frozen tissues for the DD analysis. Histologically defined matched normal and tumor tissues were initially quantified for the presence of epithelial cells by using hematoxylin- and eosin-stained slides. Genomic DNA-free total RNA was isolated from the enriched pool of cells derived from normal and tumor tissue sections, and the epithelial nature of the RNA source was confirmed further by using human cytokeratin-18 expression in reverse transcription–PCR (RT-PCR) assays. To analyze PCGEM1 expression in CaP specimens, hematoxylin- and eosin-stained slides corresponding to frozen tissue sections were used as a template, and subsequent unstained sections were superimposed on it. The microscopically defined area of the tumor cells was excised with a razor blade and used for RNA extraction by using the RNAzol B method (Tel-Test, Friendswood, TX). Poly(A)+ RNA from different cell lines was prepared by a Fast Track kit (Invitrogen). DD analysis (14) was performed with the use of a Hieroglyph mRNA profile kit (Genomyx, Foster City, CA).
Isolation of Full-Length PCGEM1 cDNA and Sequence Determination.
The full-length cDNA sequence was assembled by 5′ and 3′ rapid amplification of cDNA ends method with a Marathon-ready cDNA kit (CLONTECH). lasergene and macvector DNA analysis softwares were used to analyze DNA sequences and to define ORF regions. To obtain full-length cDNA clones, a normal prostate cDNA library (CLONTECH) was screened with a 530-bp α32P-dCTP-labeled PCGEM1 cDNA fragment, and the positive clones were characterized by DNA sequencing with an automated Applied Biosystems 310 sequencer and a dRhodamine cycle sequencing kit (PE-Applied Biosystems). In addition to sequence determination of positive clones from the library, the PCGEM1 cDNA sequence was also determined by using cDNA from the LNCaP cell line.
Chromosomal Mapping of PCGEM1 Gene by Fluorescence in Situ Hybridization.
A bacterial artificial chromosome clone containing the PCGEM1 genomic sequence was isolated (Genome Systems, St. Louis). PCGEM1 bacterial artificial chromosome clone DNA was nick translated by using spectrum orange (Vysis, Downers Grove, IL) as a direct label, and fluorescence in situ hybridization was performed with this probe on normal human male metaphase chromosome spreads (15). 4′,6-Diamidino-2-phenylindole counterstaining was carried out, and chromosomal localization was determined based on the G band analysis of inverted 4′,6-diamidino-2-phenylindole images (16). NU 200 image-acquisition and registration software was used to create the digital images. More than 20 metaphases were analyzed.
Analysis of Tissue-Specific Expression of PCGEM1 with Multiple Tissue Northern Blots.
Multiple tissue Northern blots (CLONTECH) were hybridized with the α32P-dCTP-labeled 530-bp PCGEM1 cDNA fragment and other known prostate-specific genes, which include PSA (77-mer oligo probe), PSMA (234-bp fragment from PCR product), and NKX3.1 (210-bp cDNA). As an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (17) was also used to hybridize all of the blots.
Analysis of PCGEM1 Expression in Primary CaP and CaP Cell Lines.
DNase-treated RNAs from normal and CaP tissues were used for the RT-PCR assays. The PCR conditions were optimized to be within the logarithmic phase of amplification for all of the primers used. Epithelial-cell-specific cytokeratin-18 expression and GAPDH expression were used as internal controls. PCR was performed with Amplitaq Gold from Perkin–Elmer. PCR cycles were 95°C for 10 min, 1 cycle; 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, 42 cycles; and 72°C for 5 min, 1 cycle followed by 4°C soak cycle. PCGEM1 PCR primers were 5′- TGCCTCAGCCTCCCAAGTAAC-3′ (sense) and 5′-GGCCAAAATAAAACCAAACAT-3′ (antisense). Cytokeratin-18 PCR primers were 5′-AGCGCCAGGCCCAGGAGTATGAGG-3′ (sense) and 5′-TATCCGGCGGGTGGTGGTCTTTTG-3′ (antisense), and GAPDH PCR primers were 5′-GGGGAGCCAAAAGGGTCATCATCT-3′ (sense) and 5′-GAGGGGCCATCCACAGTCTTCT-3′ (antisense). Cytokeratin-18 expression and PCGEM1 expression were analyzed at 35 and 42 cycles, respectively. PCGEM1 expression was evaluated in different CaP cell lines by RT-PCR as described above and by Northern blot hybridization with poly(A)+ RNA (17).
In Situ Hybridization.
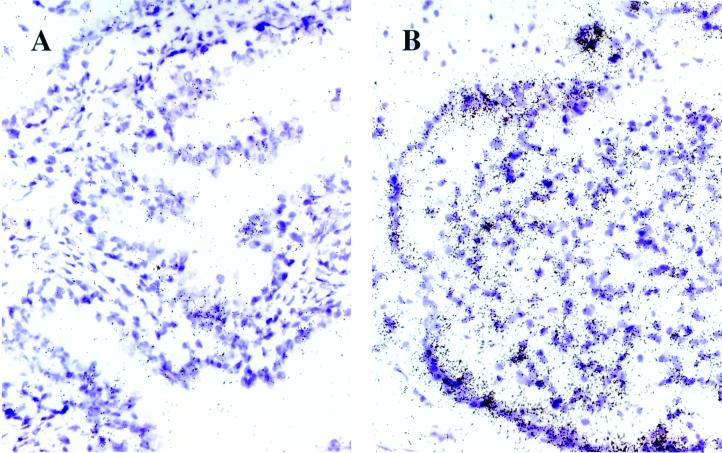
In situ hybridization was performed essentially as described by Wilkinson and Green (18). Briefly, Optimal Cutting Temperature Compound-embedded tissue slides stored at −80°C were fixed in 4% (vol/vol) paraformaldehyde, digested with proteinase K, and then fixed again in 4% (vol/vol) paraformaldehyde. After washing in PBS, sections were treated with 0.25% acetic anhydride in 0.1 M triethanolamine, washed again in PBS, and dehydrated in a graded ethanol series. Sections were hybridized with 35S-labeled riboprobes at 52°C overnight. After washing and RNase A treatment, sections were dehydrated, dipped into NTB-2 emulsion, and exposed for 11 days at 4°C. After development, slides were lightly stained with hematoxylin and mounted for microscopy. In each section, PCGEM1 expression was scored as percentage of cells showing 35S signal: 1+, 1–25%; 2+, 25–50%; 3+, 50–75%, or 4+, 75–100% (an example of a 2+ score is shown in Fig. 6B).
Figure 6.
PCGEM1 expression in normal and tumor areas of CaP tissues. In situ hybridization of 35S-labeled PCGEM1 riboprobe to matched normal (A) versus tumor (B) sections of patients with CaP. The purple areas are hematoxylin-stained cell bodies; the black dots represent the PCGEM1 expression signal. The signal is background level in the normal (A) and 2+ level in the tumor (B) section. The magnification is ×40.
Androgen Regulation of PCGEM1.
LNCaP cells were cultured in RPMI medium 1640 containing 10% (vol/vol) charcoal-stripped FBS for 4 days followed by treatment with R1881, a nonmetabolizable androgen analog (DuPont) for 12 h and 24 h at 0.1 nM and 10 nM concentrations. Poly(A)+ RNA was prepared from R1881-treated and untreated cells and was used in RT-PCR and Northern blot assays.
Results
Isolation and Characterization of the PCGEM1 cDNA.
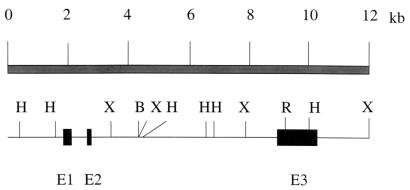
PCGEM1 was isolated from a DD analysis of paired normal and tumor tissues of CaP patients. The full length of PCGEM1 cDNA (accession no. AF223389) was obtained by 5′ and 3′ rapid amplification of cDNA ends/PCR from the original 530-bp DD product by using cDNA from normal prostate. The rapid amplification of cDNA ends/PCR products were sequenced directly. We also used the 530-bp DD fragment as a probe to screen a normal prostate cDNA library. Three overlapping cDNA clones were identified. The longest cDNA clone was 1,643 nucleotides in length with a potential polyadenylation site, ATTAAA, close to the 3′ end followed by a poly(A) tail. A full-length genomic clone was isolated and sequenced (Z.Z., unpublished work). Comparison of the cDNA and genomic sequences revealed the organization of the PCGEM1 transcription unit from three exons (Fig. 1). The GenBank database searches with blast programs did not reveal any significant homology of PCGEM1 cDNA to a previously defined gene or protein. A recent search of the high-throughput genome sequence database revealed perfect homology of PCGEM1 to a chromosome-2-derived, uncharacterized, unfinished genomic sequence (accession no. AC013401). DNA sequence analysis of the PCGEM1 cDNA (Fig. 1) and PCGEM1 genomic DNA (Z.Z., unpublished work) did not reveal a significant long ORF in either strand. The longest ORF in the sense strand was 105 nucleotides () encoding a 35-aa peptide. However, the ATG was not in a strong context of initiation.
Figure 1.
Structure of the PCGEM1 transcription unit. Sequence comparison of the isolated cDNA and genomic DNA clones revealed that the PCGEM1 gene consists of three exons. kb, kilobase; E, exon; B, BamHI; H, HindIII; X, XbaI; R, EcoRI.
Evaluation of the Coding Capacity of PCGEM1.
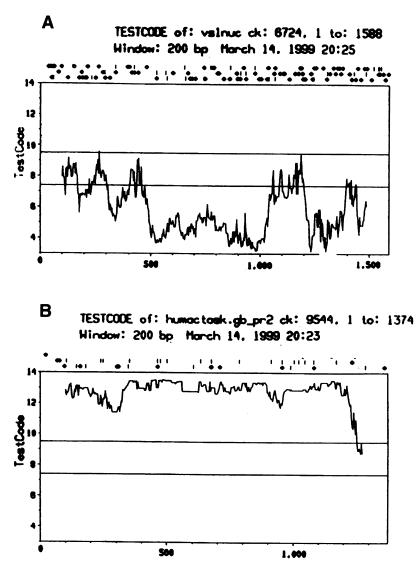
The testcode program (GCG) identifies potential protein coding sequences of longer than 200 bases by measuring the nonrandomness of the composition at every third base, independently from the reading frames. Analysis of the PCGEM1 cDNA sequence revealed that, at greater than 95% confidence level, the sequence does not contain any region with protein-coding capacity (Fig. 2A). Similar results were obtained when various published noncoding RNA sequences were analyzed with the testcode program (data not shown), whereas known protein coding regions of similar size—i.e., alpha actin (Fig. 2B)—can be detected with high fidelity. The codon preference program (GCG), which locates protein-coding regions in a reading-frame-specific manner, further suggested the absence of protein-coding capacity in the PCGEM1 gene (see www.cpdr.org). In vitro transcription/translation of PCGEM1 cDNA did not produce a detectable protein/peptide. Although we cannot unequivocally rule out whether PCGEM1 codes for a short unstable peptide, at this time, both experimental and computational approaches strongly suggest that PCGEM1 cDNA does not have protein-coding capacity.
Figure 2.
Evaluation of the coding capacity of PCGEM1. Evaluation of the coding capacity of the PCGEM1 (A) and the human alpha actin (B), independently from the reading frame, by using the TESTCODE program (GCG). The number of base pairs is indicated on the x axis, the TESTCODE values are shown on the y axis. Regions of longer than 200 base pairs above the upper line (at 9.5 value) are considered coding; those under the lower line (at 7.3 value) are considered noncoding at a confidence level greater than 95%.
Chromosomal Mapping of PCGEM1.
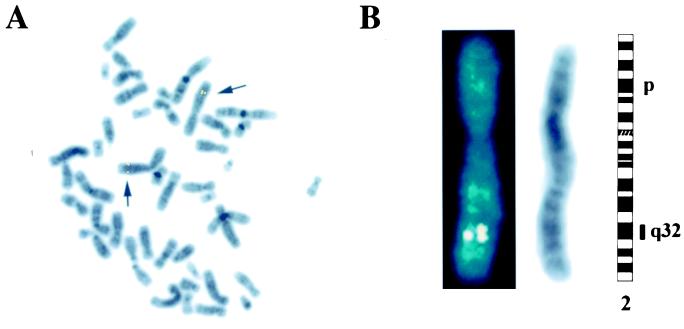
To determine the chromosomal location of the PCGEM1 gene, we used a 200-kilobase bacterial artificial chromosome clone containing PCGEM1 gene as a probe for fluorescence in situ hybridization analysis. Consistent doublet signal was observed on both homologs of chromosome 2 in every cell. Further Giemsa (G) banding pattern confirmed that PCGEM1 gene was mapped to 2q32 (Fig. 3 A and B). These results are supported further by the perfect homology of the PCGEM1 cDNA sequence to a recently described chromosome-2-derived genomic sequence (accession no. AC013401).
Figure 3.
Chromosomal localization of PCGEM1. PCGEM1 was mapped by fluorescence in situ hybridization to 2q32 by using a bacterial artificial chromosome clone. (A) A representative metaphase showing doublet signal on both homologs of chromosome 2 (arrows). (B) A 4′,6-diamidino-2-phenylindole-counterstained chromosome 2 showing the signal (Left). An inverted 4′,6-diamidino-2-phenylindole-stained chromosome 2 shown as G bands (Center). An ideogram of chromosome 2 showing the localization of the signal to band 2q32 (Right).
PCGEM1 Expression is Prostate-Tissue Specific.
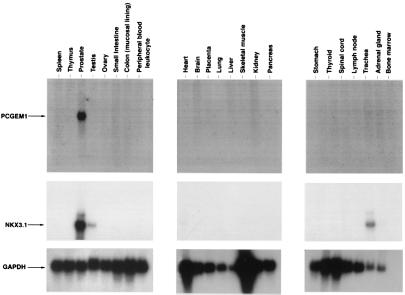
The distribution of PCGEM1 mRNA in normal human tissues was examined by Northern blot analysis. Of the 24 different human tissue mRNA analyzed (heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, colon, peripheral blood, stomach, thyroid, spinal cord, lymph node, trachea, adrenal gland, and bone marrow), a 1.7-kilobase mRNA transcript specifically hybridizing to the PCGEM1 cDNA was detected only in prostate tissue (Fig. 4). Two independent experiments revealed identical results. Further analysis of RNA Master blot (CLONTECH) containing mRNA from 50 different tissues from human adult and fetal tissues, yeast RNA, and Escherichia coli RNA confirmed the prostate-tissue specificity of PCGEM1 gene (see www.cpdr.org). Northern blot analysis revealed that the prostate-tissue specificity of PCGEM1 was comparable to the well known prostate marker PSA and superior to other prostate-specific genes: NKX3.1 (Fig. 4), PSMA (data not shown), prostate stem-cell antigen (8), and prostase (9). However, PCGEM1 RNA level in prostate tissue was significantly less in comparison to PSA, PSMA, and NKX3.1.
Figure 4.
Tissue-specific expression of PCGEM1. Multiple tissue Northern blots (CLONTECH) were hybridized with PCGEM1, NKX3.1, and GAPDH cDNA probe. Blots were exposed to x-ray films for different times: PCGEM1 for 48 h, NKX3.1 for 24 h, and GAPDH for 15 min.
Evaluation of PCGEM1 Expression in CaP Cell Lines and in Primary CaP.
PCGEM1 gene expression was evaluated in established CaP cell lines including LNCaP, DU145, PC3, DuPro1, and CPDR-1. In RT-PCR assays, the PCGEM1 expression was easily detectable in the widely studied androgen-responsive CaP cell line LNCaP (see www.cpdr.org). However, PCGEM1 expression was not detected in CaP cell lines DU145, PC3, and DuPro-1, all of which lack detectable levels of the androgen receptor.
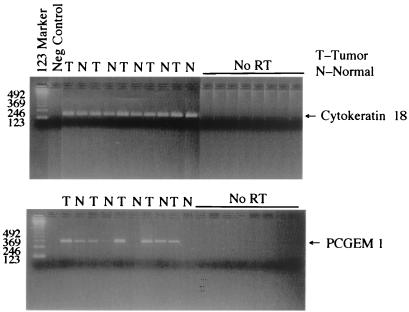
RT-PCR analysis of microdissected matched normal and tumor-tissue-derived RNAs from 23 patients with CaP revealed tumor-associated overexpression of PCGEM1 in 13 (56%) of the patients (Fig. 5). Of 23 patients, 6 (26%) did not exhibit detectable PCGEM1 expression in either normal or tumor-tissue-derived RNAs. Of 23 tumor specimens, 3 (13%) showed reduced expression in tumors. One of the patients did not exhibit any change. Expression of housekeeping gene human cytokeratin-18 or GAPDH remained constant in tumor and normal specimens of all of the patients (Fig. 5). These results were confirmed further by another set of PCGEM1-specific primers (data not shown). Importantly, RT-PCR analysis of a panel of normal and tumor tissues confirmed tumor-associated PCGEM1 overexpression originally observed in the DD experiment.
Figure 5.
Evaluation of PCGEM1 expression in primary CaP. Genomic DNA-free RNA (100-ng) samples from microdissected tissues were used to analyze expression of PCGEM1 and cytokeratin-18 by RT-PCR. Three independent experiments showed the same results.
As a complementary approach, in situ hybridization was performed to analyze PCGEM1 expression in tissue samples. Paired normal (benign) and tumor specimens from 13 additional patients were tested (representative example in Fig. 6). In 11 cases (84%), tumor-associated elevation of PCGEM1 expression was detected. In 5 of these 11 patients, the expression of PCGEM1 increased to 1+ in the tumor area from an essentially undetectable level in the normal area, on a 0 to 4+ scale (see Materials and Methods). Tumor specimens from 4 of 11 patients scored between 2+ (as shown in Fig. 6) and 4+. Of 11 patients, 2 showed focal signals with a 3+ score in the tumor area, and 1 of these patients had similar focal signal (2+) in an area pathologically designated as benign. In the remaining 2 of the 13 cases, there was no detectable signal in any of the tissue areas tested. Representative in situ hybridization photographs are available on our web site (www.cpdr.org). PCGEM1 expression seems to be restricted to glandular epithelial cells in normal and tumor specimens, and future analysis by in situ methods of higher resolution will define cell-type specificity of the PCGEM1 expression in prostate gland.
Regulation of PCGEM1 Expression by Androgens.
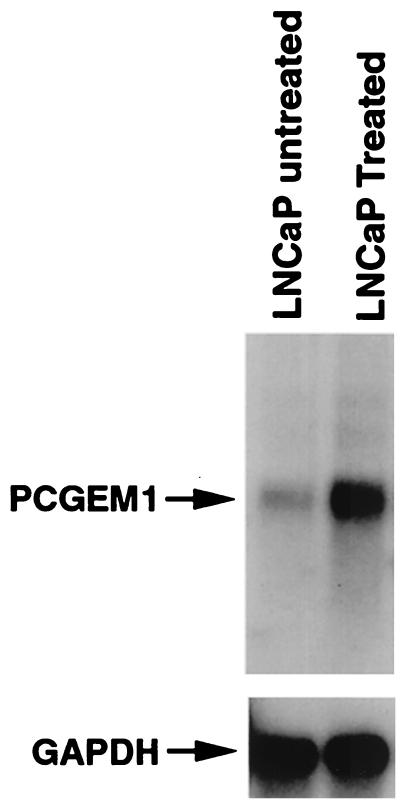
Prostate-tissue specificity of PCGEM1 expression prompted us to evaluate further whether PCGEM1 expression was regulated by androgen, the hormone that plays a critical role in prostate growth and differentiation. Northern blot analysis of LNCaP cells treated for 24 h with 10 nM synthetic androgen, R1881, showed significant induction of PCGEM1 expression compared with untreated samples (Fig. 7). PCGEM1 expression was also evaluated by RT-PCR after 0.1 nM and 10 nM R1881 treatment. R1881 induced PCGEM1 expression in a dose-dependent manner (see www.cpdr.org).
Figure 7.
Androgen regulation of PCGEM1. LNCaP cells were cultured in RPMI medium 1640 containing 10% (vol/vol) charcoal-stripped FBS for 4 days and were followed by treatment with synthetic androgen (R1881 for 24 h at 10 nanomolar concentrations). Poly(A)+ RNA from treated and untreated cells was analyzed for PCGEM1 expression by Northern blot hybridization as described for Fig. 4.
Discussion
Herein, we report the identification of an androgen-regulated prostate-specific gene, PCGEM1, which exhibits overexpression in a significant percentage of primary CaP specimens. The striking prostate-tissue specificity of PCGEM1 expression parallels the tissue specificity of PSA and has provided an impetus for a comprehensive molecular characterization of PCGEM1. Prostate-tissue-specific expression of PSA has been the key factor that led to the utility of PSA in early detection of CaP as well as in the follow-up of disease progression and minimal residual disease after treatment (2). The discovery of genes like PSMA (4), NKX3.1 (6), prostate stem-cell antigen (8), prostase (9), DD3 (11), and now PCGEM1 provides a panel of prostate-specific genes that may have potential to improve the diagnostic/prognostic capability of PSA. Relative expression levels of PSA, PSMA, NKX3.1, and PCGEM1 suggest a low abundance of PCGEM1 in normal prostate tissue. It is also important to note that PCGEM1 exhibited a significant tumor-associated overexpression in analysis of matched tumor/normal specimens of individual patients in both RT-PCR (56%) and in situ hybridization (84%) assays. The RT-PCR and in situ hybridization assays were performed with two different sets of patient samples. Apparent differences in the observed frequency of tumor-associated PCGEM1 expression by RT-PCR and in situ hybridization-based assays most likely reflect heterogeneity of PCGEM1 expression in normal and tumor-prostate tissues. Because tissue specimens were microdissected for RT-PCR assay, anticipated variations in tissue sampling might result in the underestimation of PCGEM1 expression. To address these issues, future experiments will analyze a larger cohort of specimens by using real time PCR and in situ hybridization assays. The variations in the levels of PCGEM1 expression in normal prostate tissue of different individuals need to be examined further. Focal expression of PCGEM1 was also noted in both normal and tumor cells. Comparison of the benign and tumor glands of the same patient generally showed weak or no PCGEM1 expression in normal cells and increased expression in tumor cells. PCGEM1 overexpression was observed in prostatic intraepithelial neoplasia foci of some specimens. Although, in our limited analysis, poorly differentiated cancer cells tended to show relatively higher expression of PCGEM1, a larger study is warranted to correlate PCGEM1 expression with the clinicopathologic features.
Prostate-tissue-specific expression of the PSA and probasin genes have provided further opportunities to use promoters of these genes for the prostate-tissue-specific targeting of genes in transgenic mice models of CaP. Moreover, PSA promoter-driven anticancer genes are being evaluated for their selective effects on CaP cells. Finally, prostate-specific expression of PSA, PSMA, and HK2 genes is being explored for the detection of CaP micrometastasis (19). Therefore, studies of PCGEM1 by similar approaches hold promising applications in both translational and basic research areas of CaP.
The most intriguing aspect of PCGEM1 characterization has been its lack of protein-coding capacity. Although we have not completely ruled out whether PCGEM1 codes for a short unstable peptide, careful sequencing of PCGEM1 cDNA (Fig. 1) and genomic clones (Z.Z., unpublished work), computational analysis of PCGEM1 sequence (Fig. 2 A and B), and in vitro transcription/translation experiments (data not shown) strongly suggest a noncoding nature of PCGEM1. It is interesting to note that an emerging group of mRNA-like noncoding RNAs are being discovered whose function and mechanisms of action remain poorly understood (20). Such RNA molecules have also been termed as “RNA riboregulators” because of their function(s) in development, differentiation, DNA damage, heat-shock responses, and tumorigenesis (21–24). In the context of tumorigenesis, the H19, His-1, and Bic genes code for functional noncoding mRNAs (24). In addition, a recently reported CaP-associated gene, DD3, also seems to exhibit a tissue-specific noncoding mRNA (11). In this regard, it is important to point out that PCGEM1 and DD3 may represent a distinct class of prostate-specific genes. The recent discovery of a steroid receptor coactivator as an mRNA, lacking protein-coding capacity, further emphasizes the role of RNA riboregulators in critical biochemical function(s) (25). Our recent preliminary results showed that PCGEM1 expression in NIH 3T3 cells caused a significant increase in size of colonies in colony-forming assay, suggesting that PCGEM1 cDNA confers cell proliferation and/or cell survival function(s) (V.S., unpublished work). Elevated expression of PCGEM1 in CaP cells may represent a gain in function favoring tumor-cell proliferation/survival. On the basis of our first characterization of the PCGEM1 gene, we propose that PCGEM1 belongs to a distinct class of prostate-tissue-specific genes with potential functions in prostate cell biology and the tumorigenesis of the prostate gland.
Acknowledgments
We thank Mrs. Anita Roundtree for preparing this manuscript, Mr. Naga Shanumgam for in vitro transcription/translations, and Drs. Harvey B. Pollard and Zoltan Szallasi for insightful discussions related to the structure of PCGEM1 cDNA. This research was supported in part by a grant from the Center for Prostate Disease Research, a program of the Henry M. Jackson Foundation for the Advancement of Military Medicine (Rockville, MD), which is funded by the U.S. Army Medical Research and Materiel Command.
Abbreviations
- CaP
prostate cancer
- PSA
prostate-specific antigen
- PSMA
prostate-specific membrane antigen
- DD
differential display
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF223389).
References
- 1.Landis S H, Murray T, Bolden S, Wingo P A. Ca Cancer J Clin. 1999;49:8–31. doi: 10.3322/canjclin.49.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Garnick M B, Fair W R. Sci Am. 1998;279(6):74–83. doi: 10.1038/scientificamerican1298-74. [DOI] [PubMed] [Google Scholar]
- 3.Augustus M, Moul J W, Srivastava S. In: Molecular Pathology of Early Cancer. Srivastava S, Henson D E, Gazden A, editors. Amsterdam: IOS; 1999. pp. 321–340. [Google Scholar]
- 4.Fair W R, Israeli R S, Heston W D. Prostate. 1997;32:140–148. doi: 10.1002/(sici)1097-0045(19970701)32:2<140::aid-pros9>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 5.Murphy G P, Tjoa B A, Simmons S J, Jarisch J, Bowes V A, Ragde H, Rogers M, Elgamal A, Kenny G M, et al. Prostate. 1999;38:73–78. doi: 10.1002/(sici)1097-0045(19990101)38:1<73::aid-pros9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 6.He W W, Sciavolino P J, Wing J, Augustus M, Hudson P, Meissner P S, Curtis R T, Schell B K, Bostwick D G, et al. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia-Gaur R, Donjacour A A, Sciavolino P J, Kim M, Desai N, Young P, Norton C R, Gridley T, Cardiff R D, Cunha G R, et al. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiter R E, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yarnashiro L, LeBeau M M, et al. Proc Natl Acad Sci USA. 1998;95:1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson P S, Gan L, Ferguson C, Moss P, Gelinas R, Hood L, Wang K. Proc Natl Acad Sci USA. 1999;96:3114–3119. doi: 10.1073/pnas.96.6.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin B, Ferguson C, White J T, Wang S, Vessella R, True L D, Hood L, Nelson P S. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 11.Bussemakers M J H, Van Bokhoven A, Verhaegh G W, Smit F P, Karthaus H F, Schalken J A, Debruyne F M, Ru N, Isaacs W B. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 12.Srikantan V, Sesterhenn I A, Davis L, Hankins G R, Avallone F A, Livezey J R, Connelly R, Mostofi F K, McLeod D G, Moul J W, et al. Int J Cancer. 1999;84:331–335. doi: 10.1002/(sici)1097-0215(19990621)84:3<331::aid-ijc23>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 13.Gingrich J R, Tucker J A, Walther P J, Day J W, Poulton S H M, Webb K S. J Urol. 1991;146:915–919. doi: 10.1016/s0022-5347(17)37960-0. [DOI] [PubMed] [Google Scholar]
- 14.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 15.Heselmeyer K, Macville M, Schrock E, Blegen H, Hellstrom A C, Shah K, Auer G, Ried T. Genes Chromosomes Cancer. 1997;4:233–240. [PubMed] [Google Scholar]
- 16.Mitelman F. In: An International System for Human Cytogenetics Nomenclature. Mitelman F, editor. Basel: Karger; 1995. [Google Scholar]
- 17.Gaddipati J P, McLeod D G, Sesterhenn I A, Hussussian C J, Tong Y A, Seth P, Dracopoli N C, Moul J W, Srivastava S. Prostate. 1997;30:188–194. doi: 10.1002/(sici)1097-0045(19970215)30:3<188::aid-pros7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson D, Green J. In: Postimplantation Mammalian Embryos. Copp A J, Cokroft D L, editors. London: Oxford Univ. Press; 1990. pp. 155–171. [Google Scholar]
- 19.de la Taille A, Olsson C A, Katz A E. Oncology. 1999;13:187–194. [PubMed] [Google Scholar]
- 20.Erdmann V A, Szymanski M, Hochberg A, de Groot N, Barciszewski J. Nucleic Acids Research. 1999;27:192–195. doi: 10.1093/nar/27.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crespi M D, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velleca M A, Wallace M C, Merlie J P. Mol Cell Biol. 1994;14:7095–7104. doi: 10.1128/mcb.14.11.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeda K, Ichijo H, Fujii M, Mochida Y, Saitoh M, Nishitoh H, Sampath T K, Miyazono K. J Biol Chem. 1998;273:17079–17085. doi: 10.1074/jbc.273.27.17079. [DOI] [PubMed] [Google Scholar]
- 24.Askew D S, Xu F. Histol Histopathol. 1999;14:235–241. doi: 10.14670/HH-14.235. [DOI] [PubMed] [Google Scholar]
- 25.Lanz R B, McKenna N J, Onate S A, Albrecht U, Wong J, Tsai S Y, Tsai M J, O'Mally B W. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Srikantan V, Sesterhenn I A, Augustus M, Dean R, Moul J W, Carter K C, Srivastava S. J Urol. 2000;63:972–979. [PubMed] [Google Scholar]